
November 2025 Unique biology Precision therapeutics Broad impact NASDAQ LISTED FHTX Exhibit 99.1

| Forward Looking Statements 2 This presentation contains forward-looking statements that are based on management's beliefs and assumptions and on information currently available to management. All statements other than statements of historical facts contained in this presentation are forward- looking statements. In some cases, you can identify forward-looking statements by terms such as “could,” “may,” “might,” “will,” “likely,” “anticipates,” “intends,” “plans,” “seeks,” “believes,” “estimates,” “expects,” “continues,” “projects” or the negative of these terms or other similar expressions, although not all forward-looking statements contain these words. Forward-looking statements include, but are not limited to, statements concerning: the potential outcomes from our collaboration agreement with Lilly; the initiation, timing, progress and results of our research and development programs and pre-clinical studies and clinical trials, including with respect to our Phase 1 dose escalation trial of FHD-909 with Lilly; our ability to advance product candidates that we may develop and to successfully complete preclinical and clinical studies; our ability to leverage our initial programs to develop additional product candidates using our Gene Traffic Control Platform®; the impact of exogeneous factors, including macroeconomic and geopolitical circumstances, on our and our collaborators’ business operations, including our research and development programs and pre-clinical studies; developments related to our competitors and our industry; our ability to expand the target populations of our programs and the availability of patients for clinical testing; our ability to obtain regulatory approval for FHD-909 and any future product candidates from the FDA and other regulatory authorities; our ability to identify and enter into future license agreements and collaborations; our ability to continue to rely on our CDMOs and CROs for our manufacturing and research needs; regulatory developments in the United States and foreign countries; our ability to attract and retain key scientific and management personnel; the scope of protection we are able to establish, maintain and enforce for intellectual property rights covering FHD-909, our future products and our Gene Traffic Control Platform; and our use of proceeds from capital-raising transactions, estimates of our expenses, capital requirements, and needs for additional financing. You should, therefore, not rely on these forward-looking statements as representing our views as of any date subsequent to the date of this presentation. Additional important factors to be considered in connection with forward-looking statements are described in the Company's filings with the Securities and Exchange Commission, including withing the section entitled "Risk Factors" in the Company's Annual Report on Form 10-K for the fiscal year ended December 31, 2024. Any forward-looking statements represent the Company’s views only as of the date of this presentation and should not be relied upon as representing its views as of any subsequent date. The Company explicitly disclaims any obligation to update any forward-looking statements. The Company’s business is subject to substantial risks and uncertainties.
| Foghorn is a Leader in Chromatin Biology, Successfully Drugging Challenging Targets 3 Targeting chromatin regulation Implicated in up to 50% of all tumors Multi-billion $ Opportunities Unlocking selectivity of previously undruggable targets First-and-Best-in-Class Approaches Innovating selective protein degradation with capabilities in induced proximity Selective Target Engagement Leveraging a proven drug development platform with expansive potential Strategic Partnership: Multiple Programs
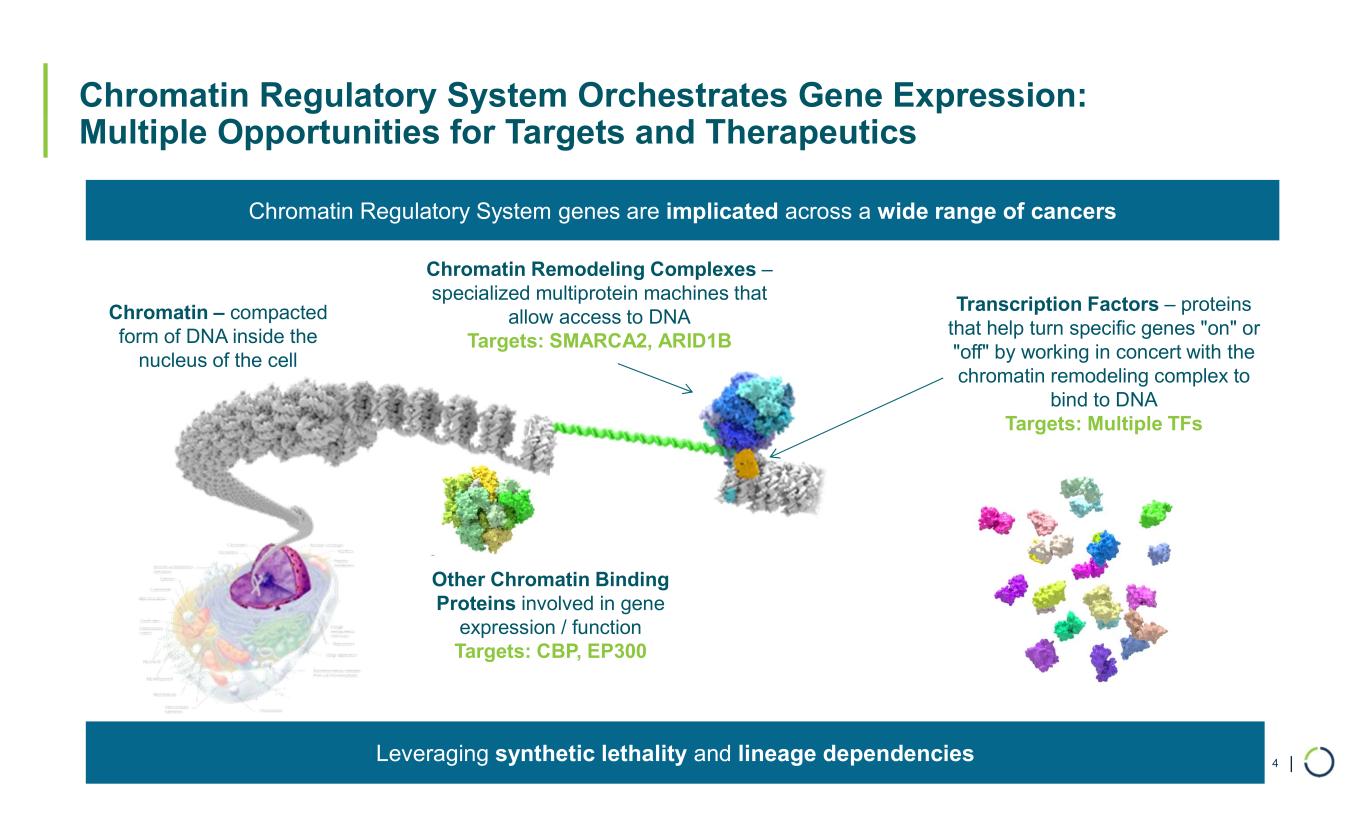
| Chromatin Regulatory System Orchestrates Gene Expression: Multiple Opportunities for Targets and Therapeutics 4 Chromatin – compacted form of DNA inside the nucleus of the cell Chromatin Remodeling Complexes – specialized multiprotein machines that allow access to DNA Targets: SMARCA2, ARID1B Transcription Factors – proteins that help turn specific genes "on" or "off" by working in concert with the chromatin remodeling complex to bind to DNA Targets: Multiple TFs Other Chromatin Binding Proteins involved in gene expression / function Targets: CBP, EP300 Chromatin Regulatory System genes are implicated across a wide range of cancers Leveraging synthetic lethality and lineage dependencies
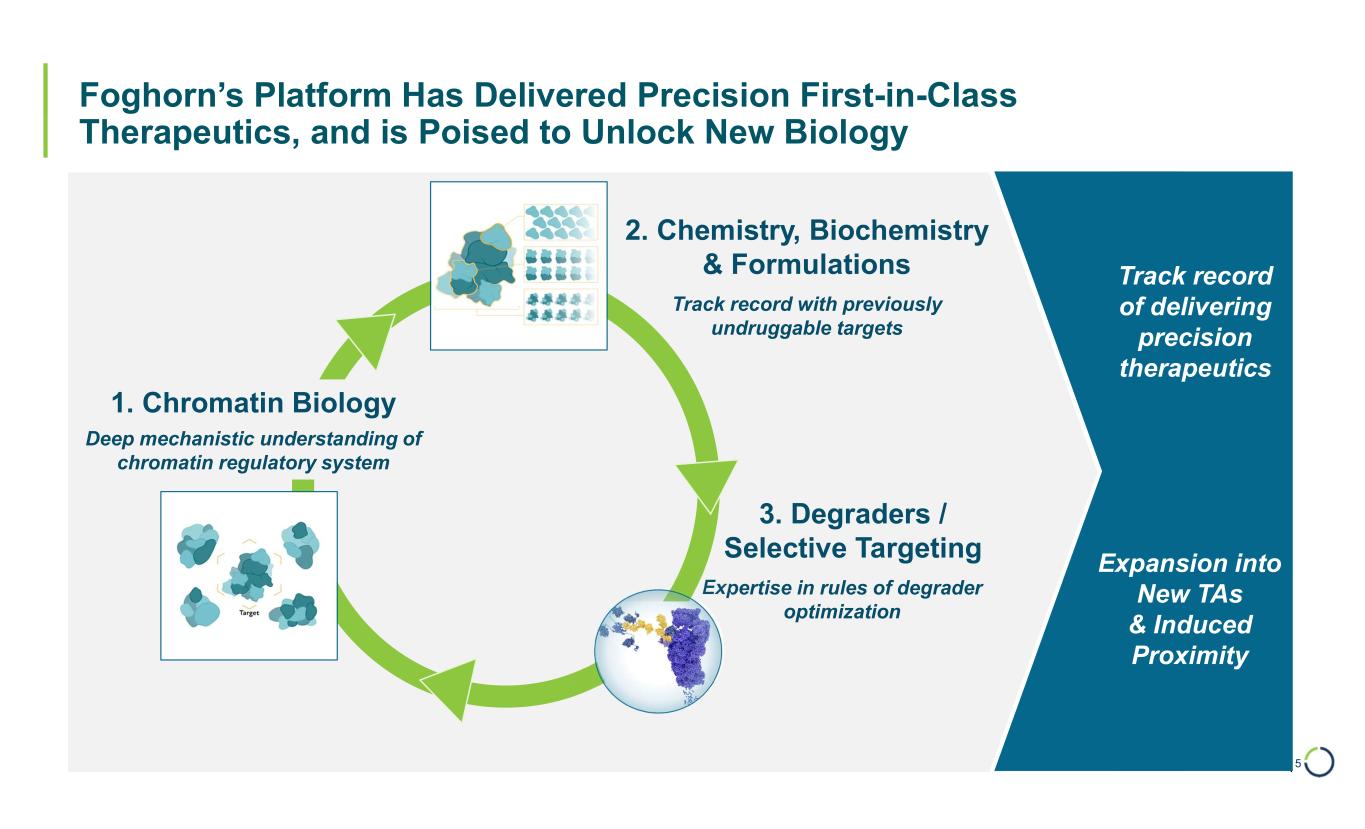
| Foghorn’s Platform Has Delivered Precision First-in-Class Therapeutics, and is Poised to Unlock New Biology 5 1. Chromatin Biology 2. Chemistry, Biochemistry & Formulations 3. Degraders / Selective Targeting Deep mechanistic understanding of chromatin regulatory system Expertise in rules of degrader optimization Track record with previously undruggable targets Expansion into New TAs & Induced Proximity Track record of delivering precision therapeutics
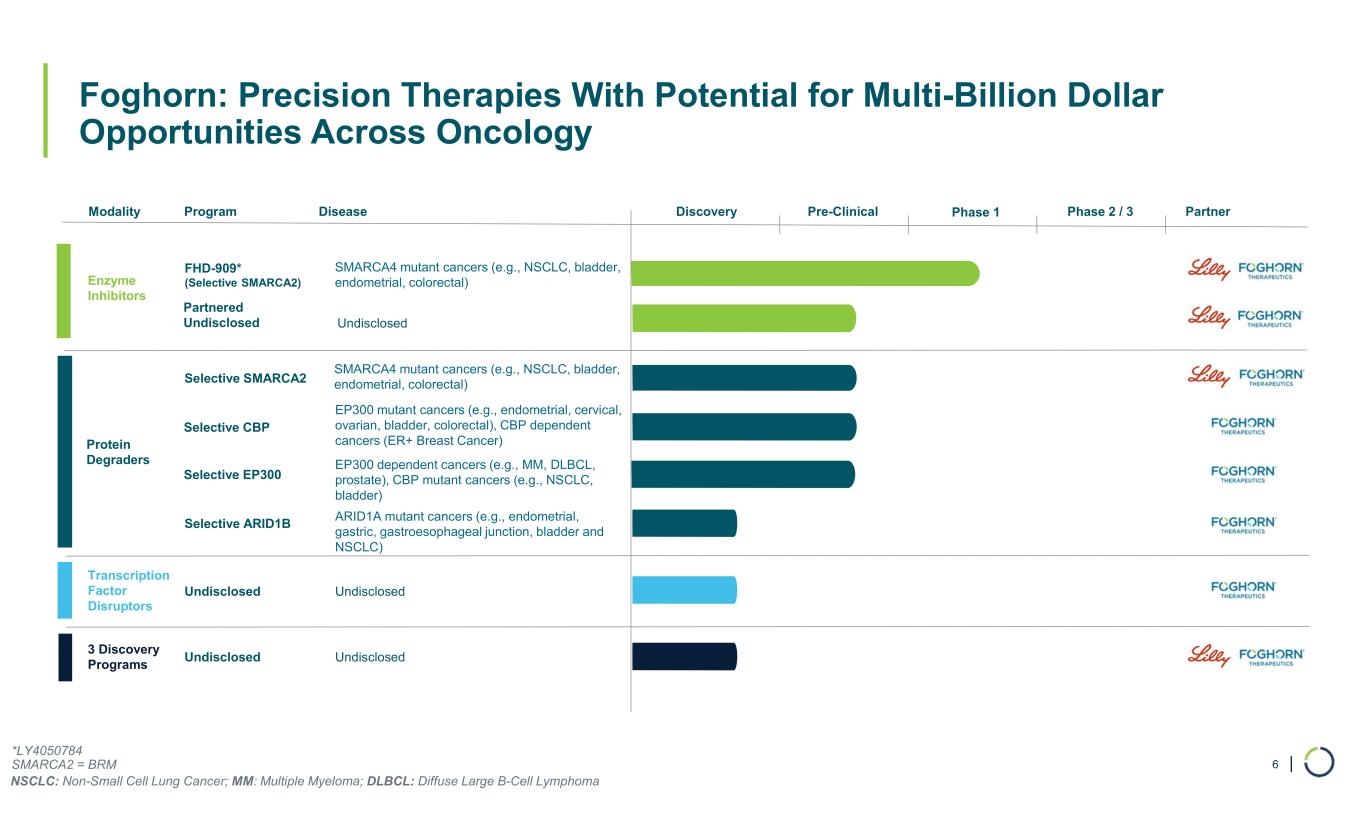
| Foghorn: Precision Therapies With Potential for Multi-Billion Dollar Opportunities Across Oncology 6 Modality Program Phase 1 Enzyme Inhibitors Transcription Factor Disruptors Undisclosed Protein Degraders Partner FHD-909* (Selective SMARCA2) 3 Discovery Programs Undisclosed Partnered Undisclosed Disease SMARCA4 mutant cancers (e.g., NSCLC, bladder, endometrial, colorectal) Selective SMARCA2 SMARCA4 mutant cancers (e.g., NSCLC, bladder, endometrial, colorectal) Selective ARID1B ARID1A mutant cancers (e.g., endometrial, gastric, gastroesophageal junction, bladder and NSCLC) Selective CBP EP300 mutant cancers (e.g., endometrial, cervical, ovarian, bladder, colorectal), CBP dependent cancers (ER+ Breast Cancer) Selective EP300 EP300 dependent cancers (e.g., MM, DLBCL, prostate), CBP mutant cancers (e.g., NSCLC, bladder) Undisclosed Undisclosed Undisclosed Pre-Clinical Phase 2 / 3Discovery *LY4050784 SMARCA2 = BRM NSCLC: Non-Small Cell Lung Cancer; MM: Multiple Myeloma; DLBCL: Diffuse Large B-Cell Lymphoma

| Advancing Multiple Programs with Near-Term Milestones 7 Phase 1 Monotherapy DataFHD-909* (LY4050784) (Selective SMARCA2 Inhibitor) 2026IND-Ready Selective CBP Degrader 2026IND-enabling StudiesSelective EP300 Degrader ConfidentialTransition to Lilly / INDSelective SMARCA2 Degrader* ConfidentialTransition to Lilly / INDLilly Target #2** 2026In Vivo Proof of ConceptSelective ARID1B Degrader * 50/50 U.S. economic split, ex-U.S. royalties. ** Pending Lilly decision to proceed, 50/50 U.S. economic split, ex-U.S. royalties. Confidential
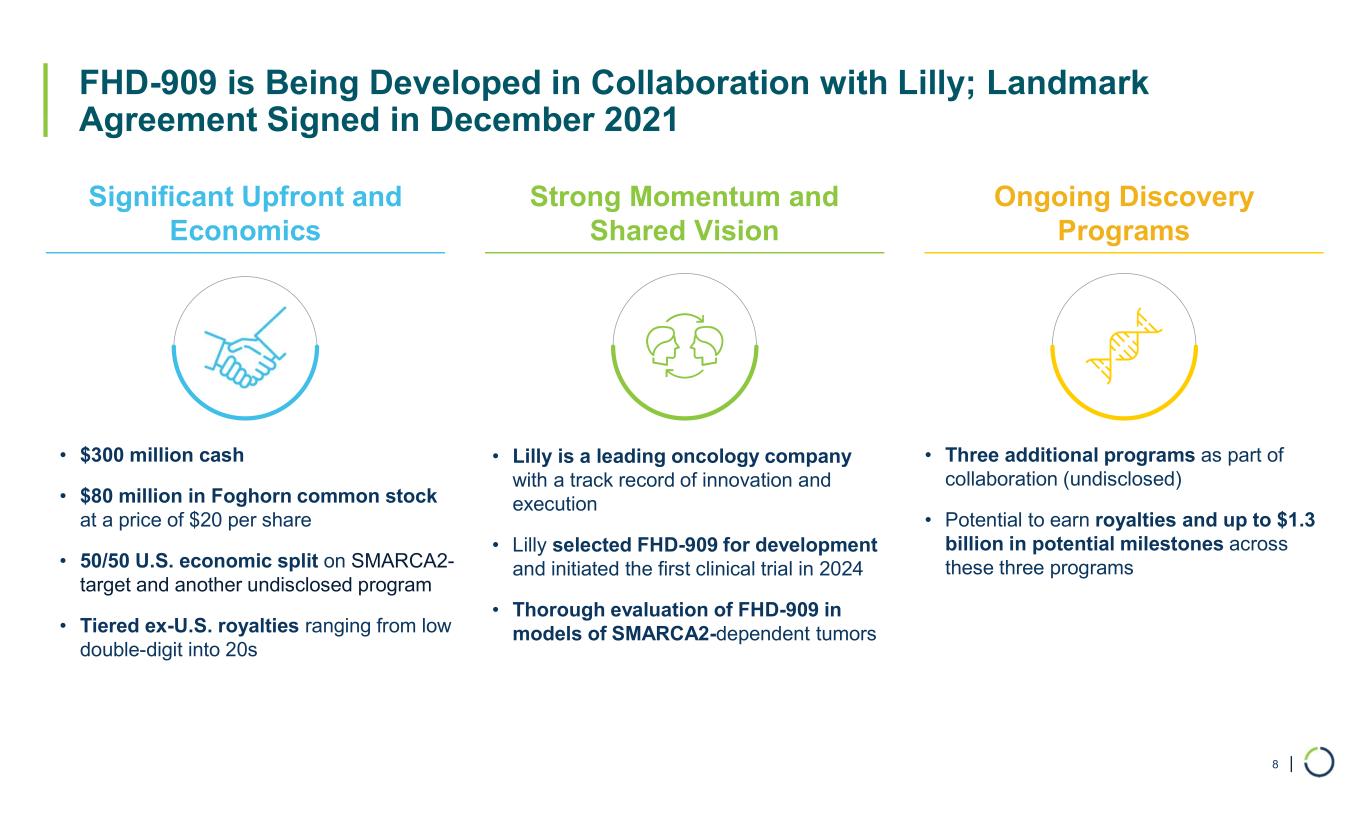
| FHD-909 is Being Developed in Collaboration with Lilly; Landmark Agreement Signed in December 2021 Significant Upfront and Economics Strong Momentum and Shared Vision Ongoing Discovery Programs • $300 million cash • $80 million in Foghorn common stock at a price of $20 per share • 50/50 U.S. economic split on SMARCA2- target and another undisclosed program • Tiered ex-U.S. royalties ranging from low double-digit into 20s • Lilly is a leading oncology company with a track record of innovation and execution • Lilly selected FHD-909 for development and initiated the first clinical trial in 2024 • Thorough evaluation of FHD-909 in models of SMARCA2-dependent tumors • Three additional programs as part of collaboration (undisclosed) • Potential to earn royalties and up to $1.3 billion in potential milestones across these three programs 8
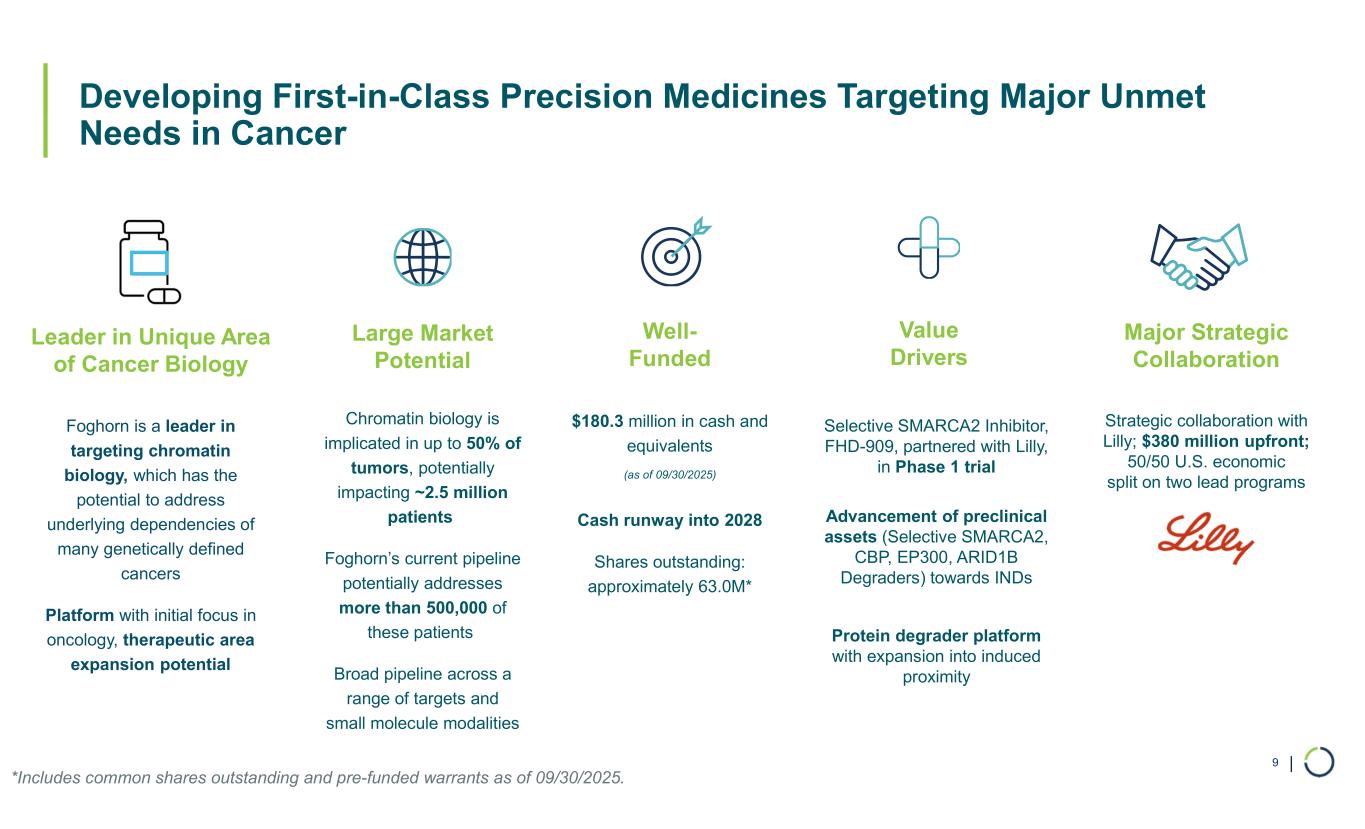
| Developing First-in-Class Precision Medicines Targeting Major Unmet Needs in Cancer 9 Large Market Potential Chromatin biology is implicated in up to 50% of tumors, potentially impacting ~2.5 million patients Foghorn’s current pipeline potentially addresses more than 500,000 of these patients Broad pipeline across a range of targets and small molecule modalities Major Strategic Collaboration Strategic collaboration with Lilly; $380 million upfront; 50/50 U.S. economic split on two lead programs Well- Funded $180.3 million in cash and equivalents (as of 09/30/2025) Cash runway into 2028 Shares outstanding: approximately 63.0M* Value Drivers Selective SMARCA2 Inhibitor, FHD-909, partnered with Lilly, in Phase 1 trial Advancement of preclinical assets (Selective SMARCA2, CBP, EP300, ARID1B Degraders) towards INDs Protein degrader platform with expansion into induced proximity Leader in Unique Area of Cancer Biology Foghorn is a leader in targeting chromatin biology, which has the potential to address underlying dependencies of many genetically defined cancers Platform with initial focus in oncology, therapeutic area expansion potential *Includes common shares outstanding and pre-funded warrants as of 09/30/2025.

Selective SMARCA2 Program For SMARCA4-Mutant Cancers • FHD-909 (LY4050784) – Selective SMARCA2 Inhibitor 10
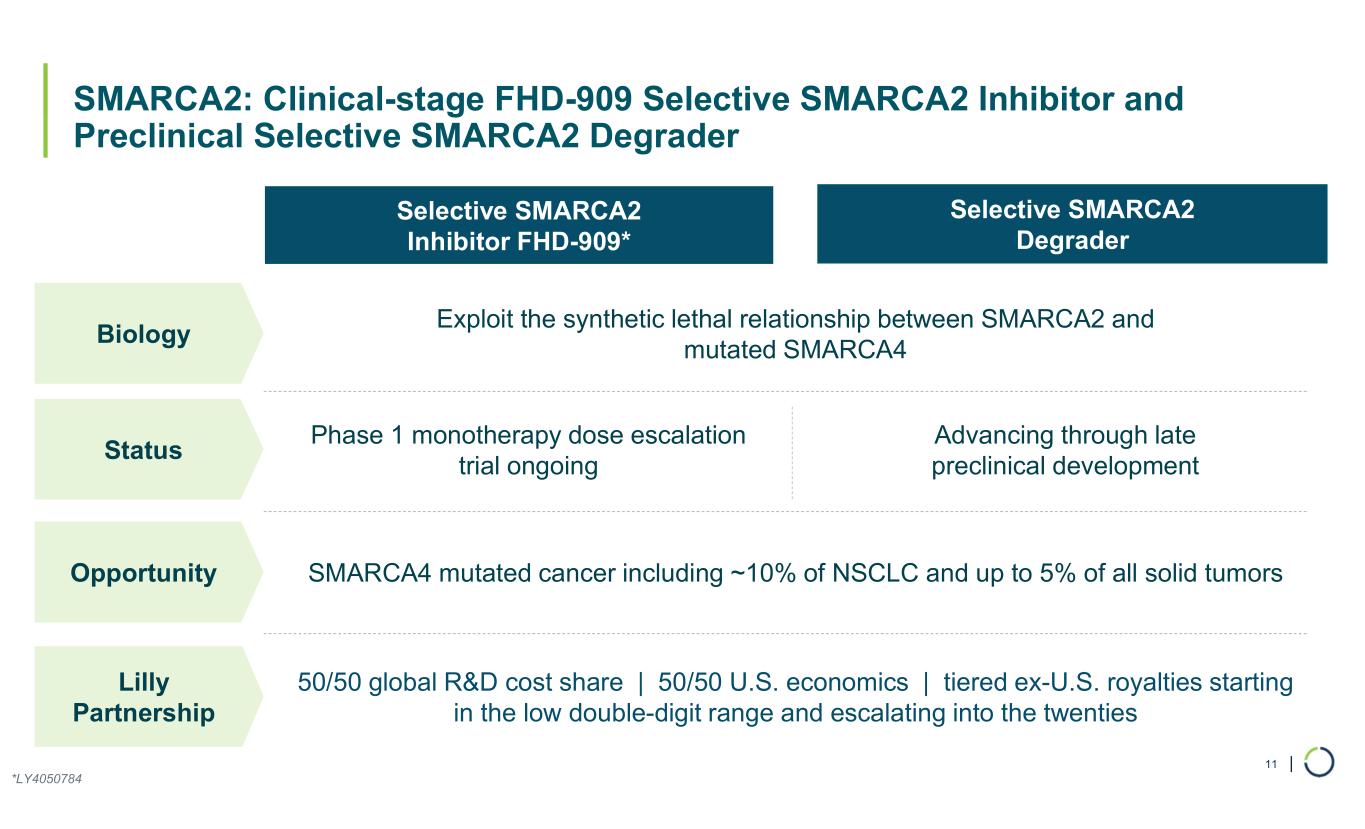
| SMARCA2: Clinical-stage FHD-909 Selective SMARCA2 Inhibitor and Preclinical Selective SMARCA2 Degrader 11 Selective SMARCA2 Inhibitor FHD-909* Selective SMARCA2 Degrader Biology Exploit the synthetic lethal relationship between SMARCA2 and mutated SMARCA4 Status Phase 1 monotherapy dose escalation trial ongoing Advancing through late preclinical development Opportunity SMARCA4 mutated cancer including ~10% of NSCLC and up to 5% of all solid tumors Lilly Partnership 50/50 global R&D cost share | 50/50 U.S. economics | tiered ex-U.S. royalties starting in the low double-digit range and escalating into the twenties *LY4050784
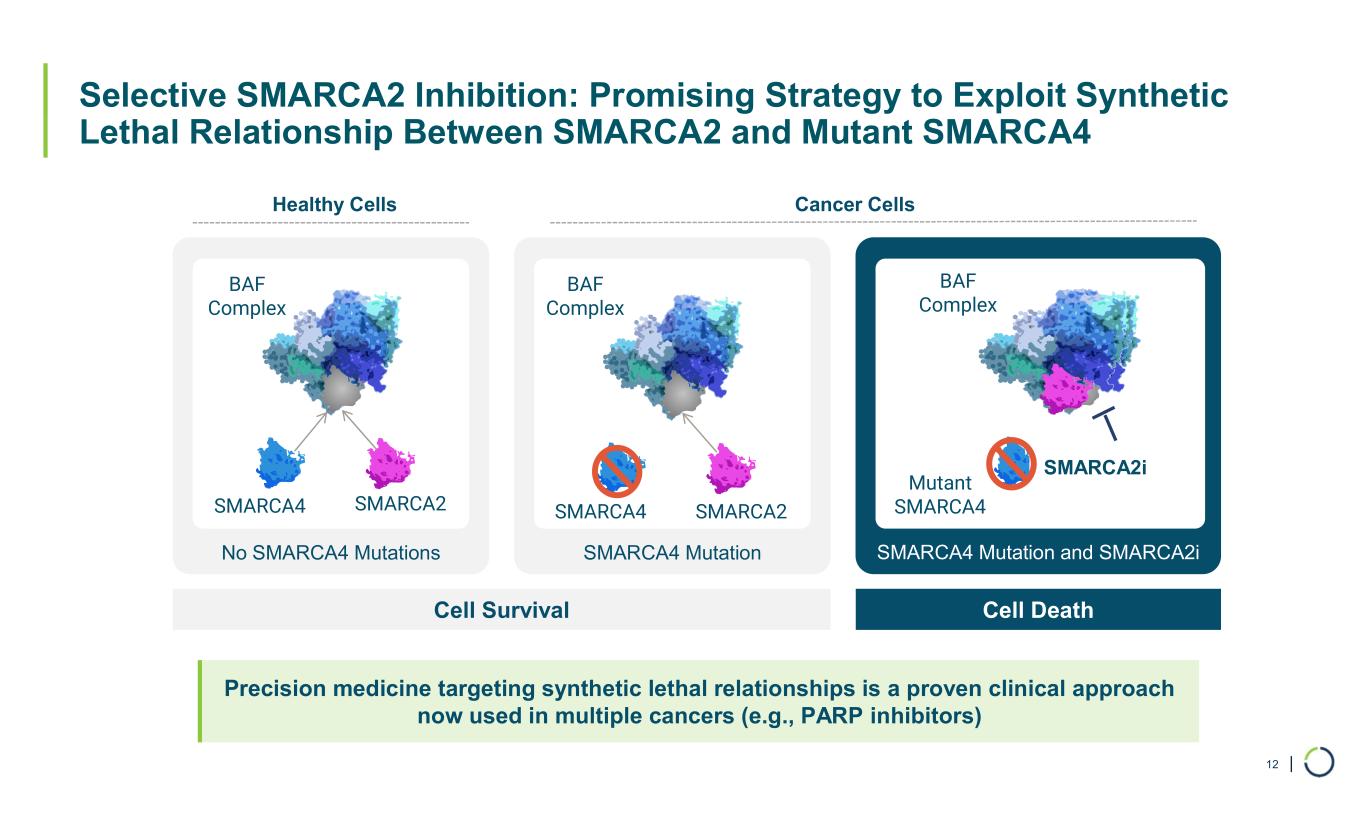
| Selective SMARCA2 Inhibition: Promising Strategy to Exploit Synthetic Lethal Relationship Between SMARCA2 and Mutant SMARCA4 12 No SMARCA4 Mutations SMARCA4 Mutation SMARCA4 Mutation and SMARCA2i Cell Survival Cell Death Healthy Cells Cancer Cells SMARCA4 SMARCA2 BAF Complex BAF Complex Mutant SMARCA4 BAF Complex SMARCA2i Precision medicine targeting synthetic lethal relationships is a proven clinical approach now used in multiple cancers (e.g., PARP inhibitors) SMARCA4 SMARCA2
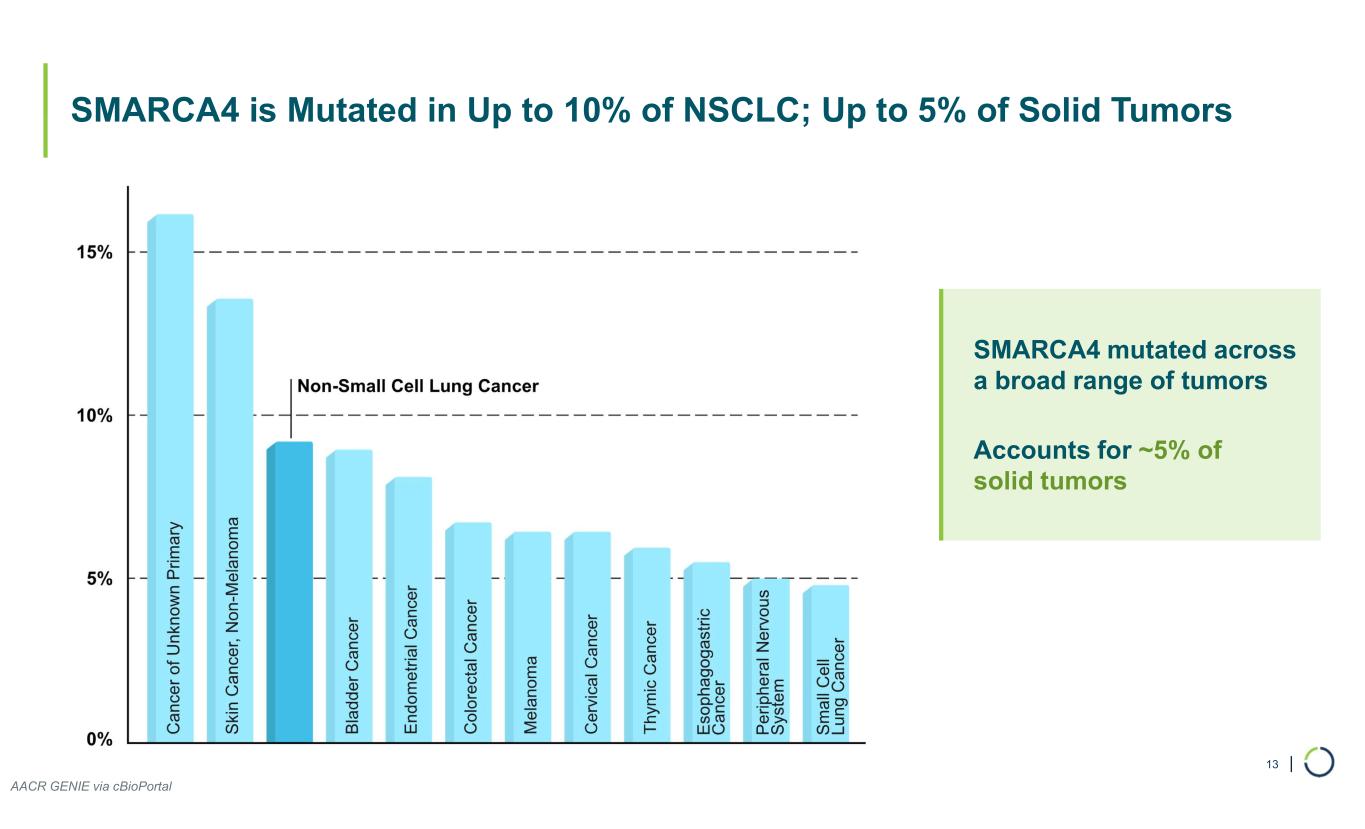
| SMARCA4 is Mutated in Up to 10% of NSCLC; Up to 5% of Solid Tumors 13 SMARCA4 mutated across a broad range of tumors Accounts for ~5% of solid tumors AACR GENIE via cBioPortal
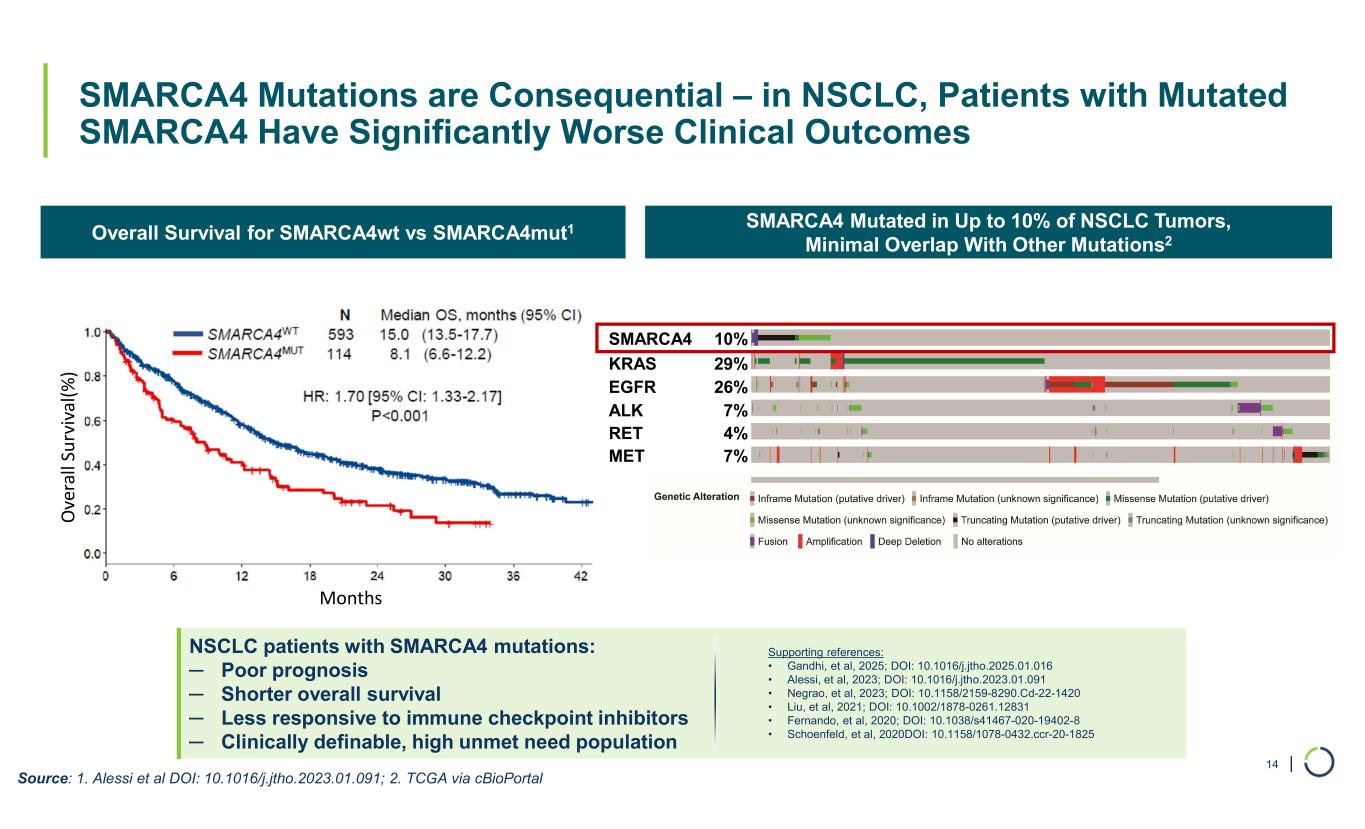
| NSCLC patients with SMARCA4 mutations: ─ Poor prognosis ─ Shorter overall survival ─ Less responsive to immune checkpoint inhibitors ─ Clinically definable, high unmet need population SMARCA4 Mutated in Up to 10% of NSCLC Tumors, Minimal Overlap With Other Mutations2Overall Survival for SMARCA4wt vs SMARCA4mut1 SMARCA4 Mutations are Consequential – in NSCLC, Patients with Mutated SMARCA4 Have Significantly Worse Clinical Outcomes 14 SMARCA4 10% KRAS 29% EGFR 26% ALK 7% RET 4% MET 7% Source: 1. Alessi et al DOI: 10.1016/j.jtho.2023.01.091; 2. TCGA via cBioPortal O ve ra ll Su rv iv al (% ) Months Supporting references: • Gandhi, et al, 2025; DOI: 10.1016/j.jtho.2025.01.016 • Alessi, et al, 2023; DOI: 10.1016/j.jtho.2023.01.091 • Negrao, et al, 2023; DOI: 10.1158/2159-8290.Cd-22-1420 • Liu, et al, 2021; DOI: 10.1002/1878-0261.12831 • Fernando, et al, 2020; DOI: 10.1038/s41467-020-19402-8 • Schoenfeld, et al, 2020DOI: 10.1158/1078-0432.ccr-20-1825
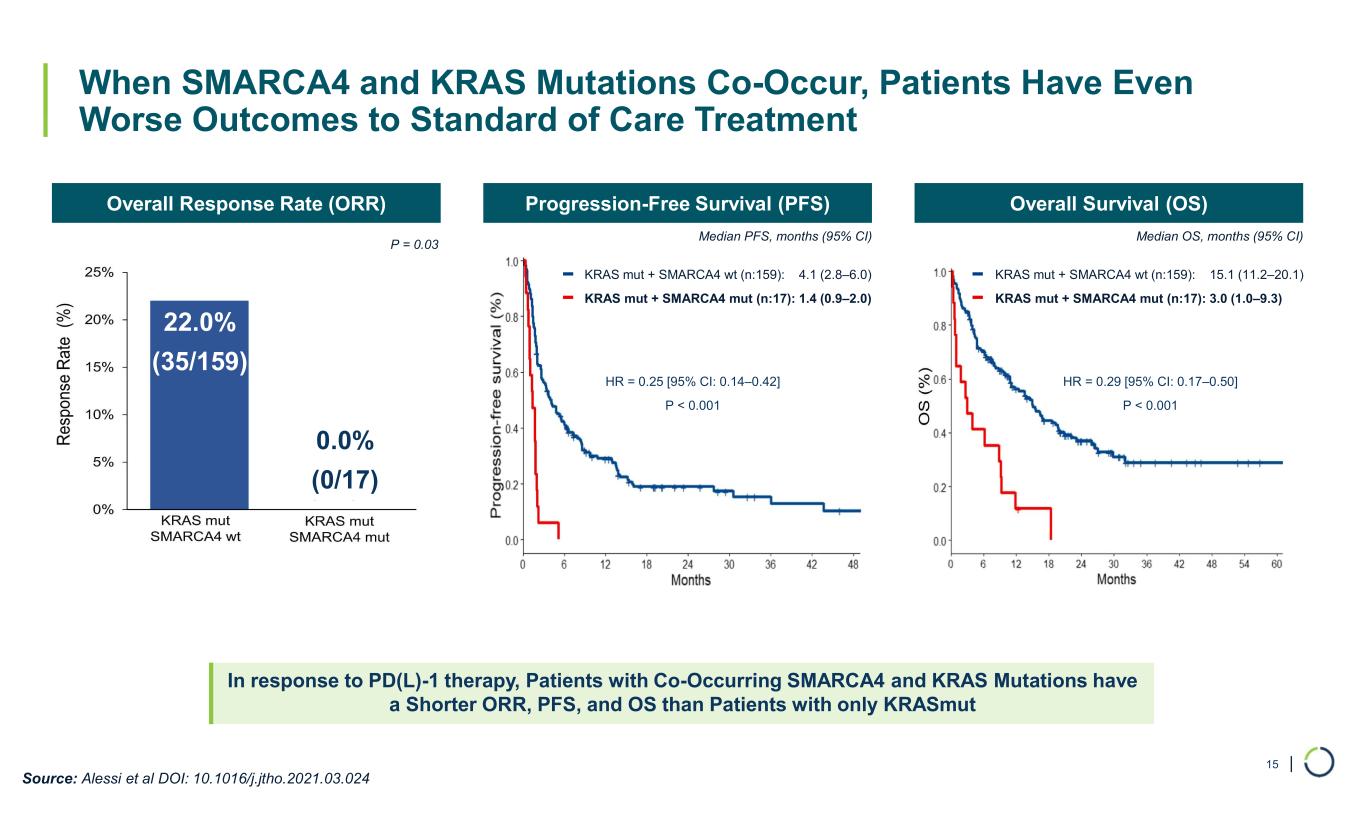
| When SMARCA4 and KRAS Mutations Co-Occur, Patients Have Even Worse Outcomes to Standard of Care Treatment In response to PD(L)-1 therapy, Patients with Co-Occurring SMARCA4 and KRAS Mutations have a Shorter ORR, PFS, and OS than Patients with only KRASmut 15 Overall Response Rate (ORR) Source: Alessi et al DOI: 10.1016/j.jtho.2021.03.024 Progression-Free Survival (PFS) Overall Survival (OS) Median PFS, months (95% CI) Median OS, months (95% CI) KRAS mut + SMARCA4 wt (n:159): 15.1 (11.2–20.1) KRAS mut + SMARCA4 mut (n:17): 3.0 (1.0–9.3) HR = 0.25 [95% CI: 0.14–0.42] P < 0.001 HR = 0.29 [95% CI: 0.17–0.50] P < 0.001 0.0% (0/17) KRAS mut + SMARCA4 wt (n:159): 4.1 (2.8–6.0) KRAS mut + SMARCA4 mut (n:17): 1.4 (0.9–2.0) P = 0.03 22.0% (35/159)
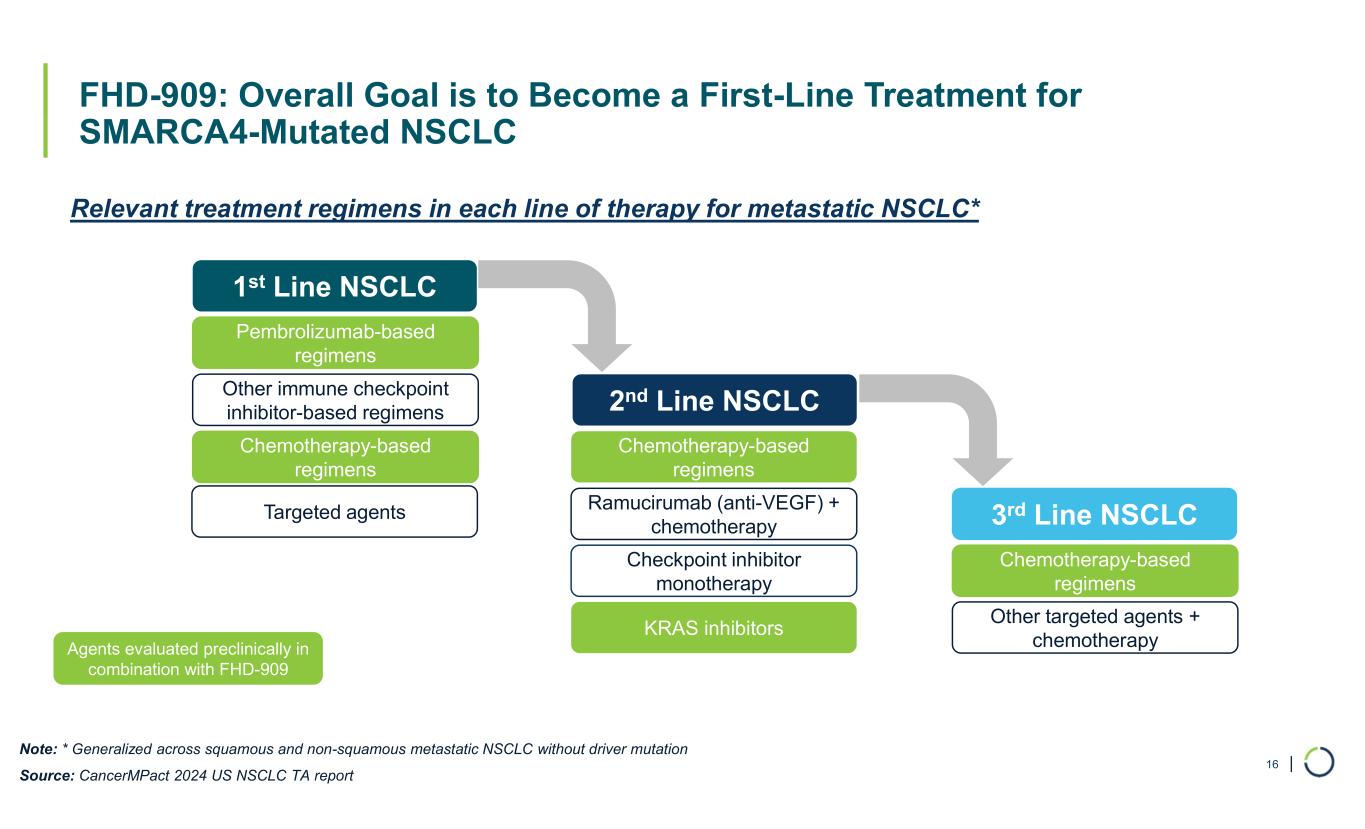
| FHD-909: Overall Goal is to Become a First-Line Treatment for SMARCA4-Mutated NSCLC 16 1st Line NSCLC Pembrolizumab-based regimens Other immune checkpoint inhibitor-based regimens Chemotherapy-based regimens 2nd Line NSCLC Chemotherapy-based regimens Ramucirumab (anti-VEGF) + chemotherapy Checkpoint inhibitor monotherapy 3rd Line NSCLC Chemotherapy-based regimens Other targeted agents + chemotherapy Relevant treatment regimens in each line of therapy for metastatic NSCLC* Agents evaluated preclinically in combination with FHD-909 KRAS inhibitors Targeted agents Note: * Generalized across squamous and non-squamous metastatic NSCLC without driver mutation Source: CancerMPact 2024 US NSCLC TA report
| Chemotherapy Targeted Therapies FHD-909 is Positioned to Be Evaluated in Combination with Standard Therapies FHD-909 shows significant anti-tumor activity preclinically in combination with standard therapies in NSCLC 17 Cisplatin PemetrexedPaclitaxel KRAS inhibitorsPembrolizumab
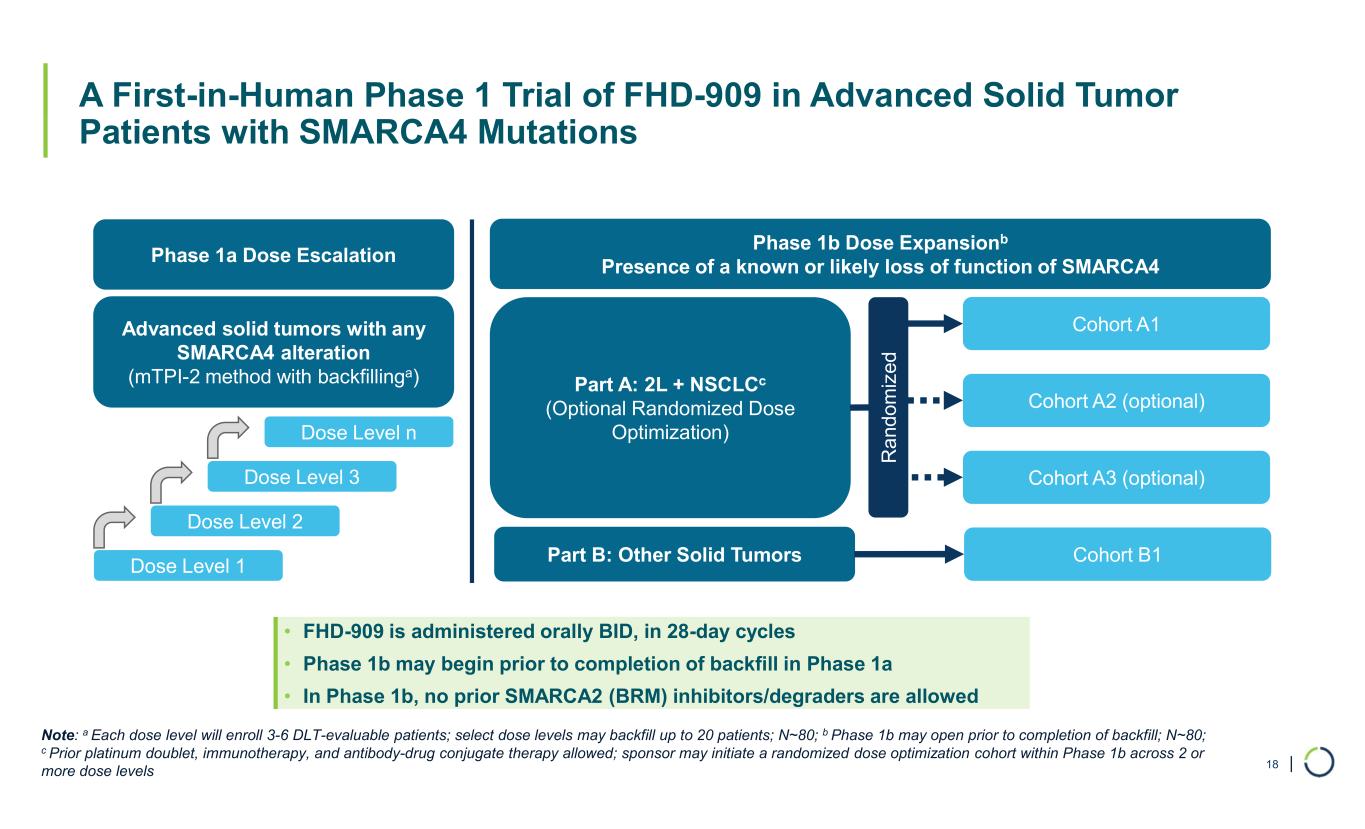
| A First-in-Human Phase 1 Trial of FHD-909 in Advanced Solid Tumor Patients with SMARCA4 Mutations Phase 1a Dose Escalation Advanced solid tumors with any SMARCA4 alteration (mTPI-2 method with backfillinga) Dose Level 1 Dose Level 2 Dose Level 3 Dose Level n Phase 1b Dose Expansionb Presence of a known or likely loss of function of SMARCA4 Part A: 2L + NSCLCc (Optional Randomized Dose Optimization) Part B: Other Solid Tumors Cohort A1 Cohort A2 (optional) Cohort A3 (optional) R a nd om iz ed Cohort B1 • FHD-909 is administered orally BID, in 28-day cycles • Phase 1b may begin prior to completion of backfill in Phase 1a • In Phase 1b, no prior SMARCA2 (BRM) inhibitors/degraders are allowed Note: a Each dose level will enroll 3-6 DLT-evaluable patients; select dose levels may backfill up to 20 patients; N~80; b Phase 1b may open prior to completion of backfill; N~80; c Prior platinum doublet, immunotherapy, and antibody-drug conjugate therapy allowed; sponsor may initiate a randomized dose optimization cohort within Phase 1b across 2 or more dose levels 18

Degrader Programs • Selective CBP Degrader • Selective EP300 Degrader • Selective ARID1B Degrader 19
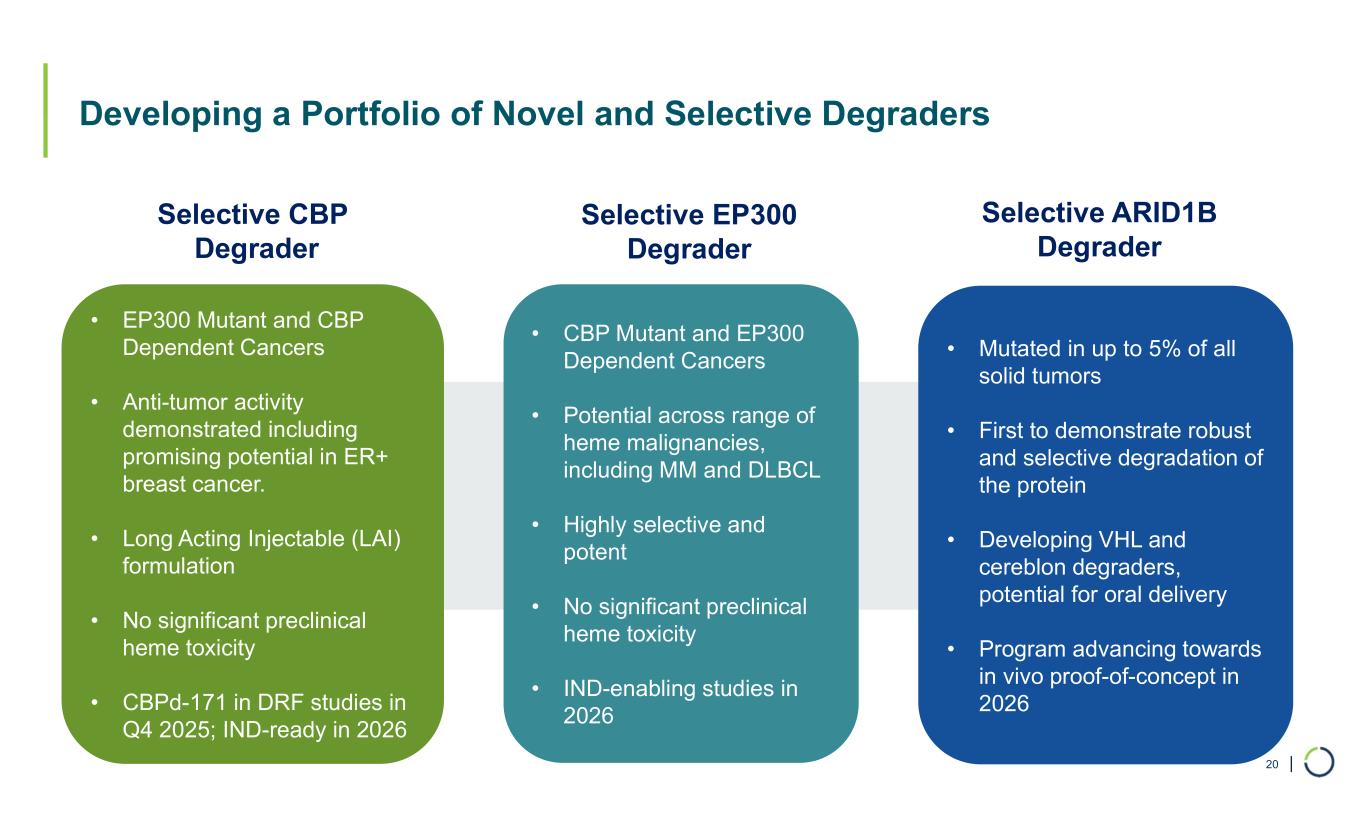
| Developing a Portfolio of Novel and Selective Degraders 20 Selective ARID1B Degrader Selective CBP Degrader • Mutated in up to 5% of all solid tumors • First to demonstrate robust and selective degradation of the protein • Developing VHL and cereblon degraders, potential for oral delivery • Program advancing towards in vivo proof-of-concept in 2026 • EP300 Mutant and CBP Dependent Cancers • Anti-tumor activity demonstrated including promising potential in ER+ breast cancer. • Long Acting Injectable (LAI) formulation • No significant preclinical heme toxicity • CBPd-171 in DRF studies in Q4 2025; IND-ready in 2026 • CBP Mutant and EP300 Dependent Cancers • Potential across range of heme malignancies, including MM and DLBCL • Highly selective and potent • No significant preclinical heme toxicity • IND-enabling studies in 2026 Selective EP300 Degrader
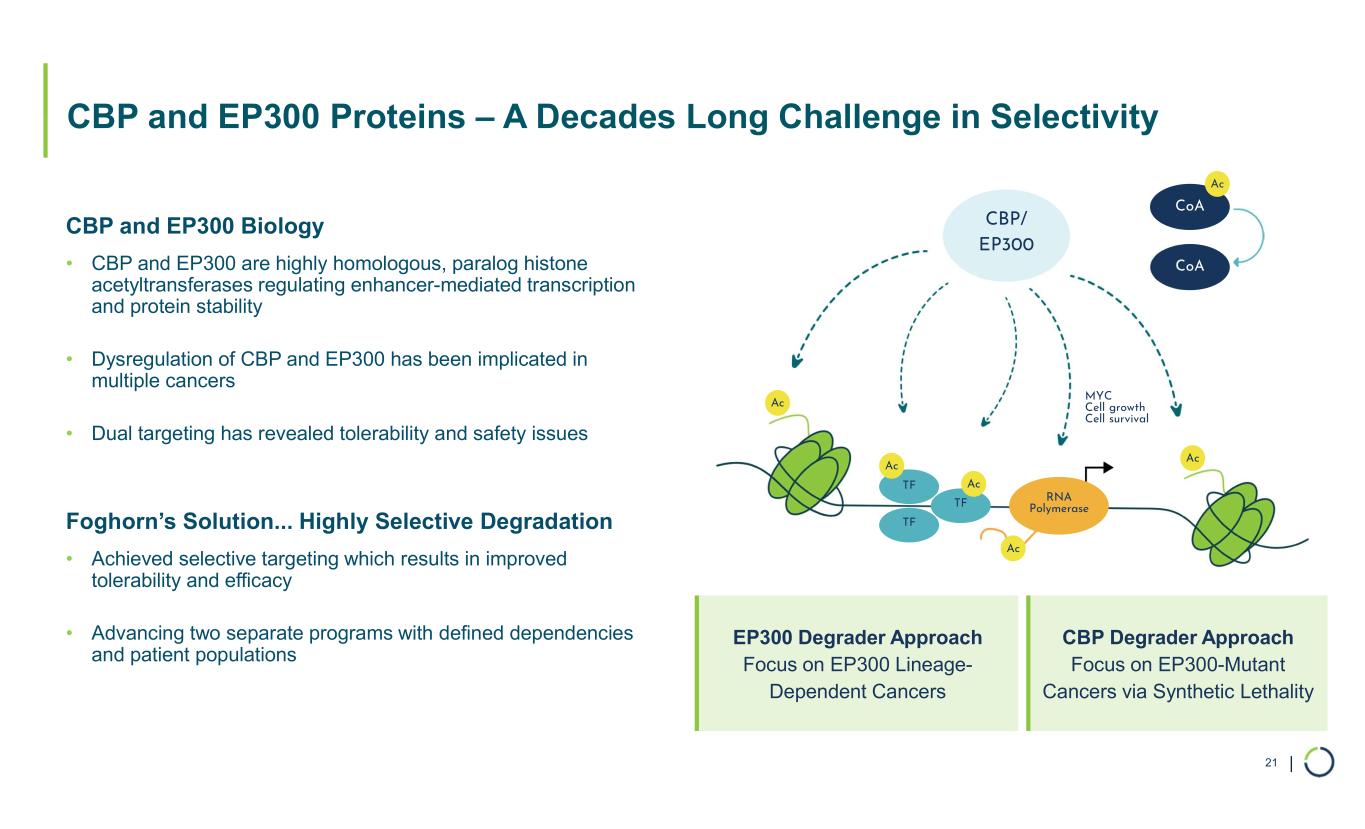
| CBP and EP300 Proteins – A Decades Long Challenge in Selectivity 21 CBP and EP300 Biology • CBP and EP300 are highly homologous, paralog histone acetyltransferases regulating enhancer-mediated transcription and protein stability • Dysregulation of CBP and EP300 has been implicated in multiple cancers • Dual targeting has revealed tolerability and safety issues Foghorn’s Solution... Highly Selective Degradation • Achieved selective targeting which results in improved tolerability and efficacy • Advancing two separate programs with defined dependencies and patient populations CoA TF TF TF RNA Polymerase Ac Ac CoA Ac Ac Ac CBP/ EP300 MYC Cell growth Cell survival Ac EP300 Degrader Approach Focus on EP300 Lineage- Dependent Cancers CBP Degrader Approach Focus on EP300-Mutant Cancers via Synthetic Lethality

Selective CBP Degrader For EP300-Mutant and CBP-Dependent Cancers 22
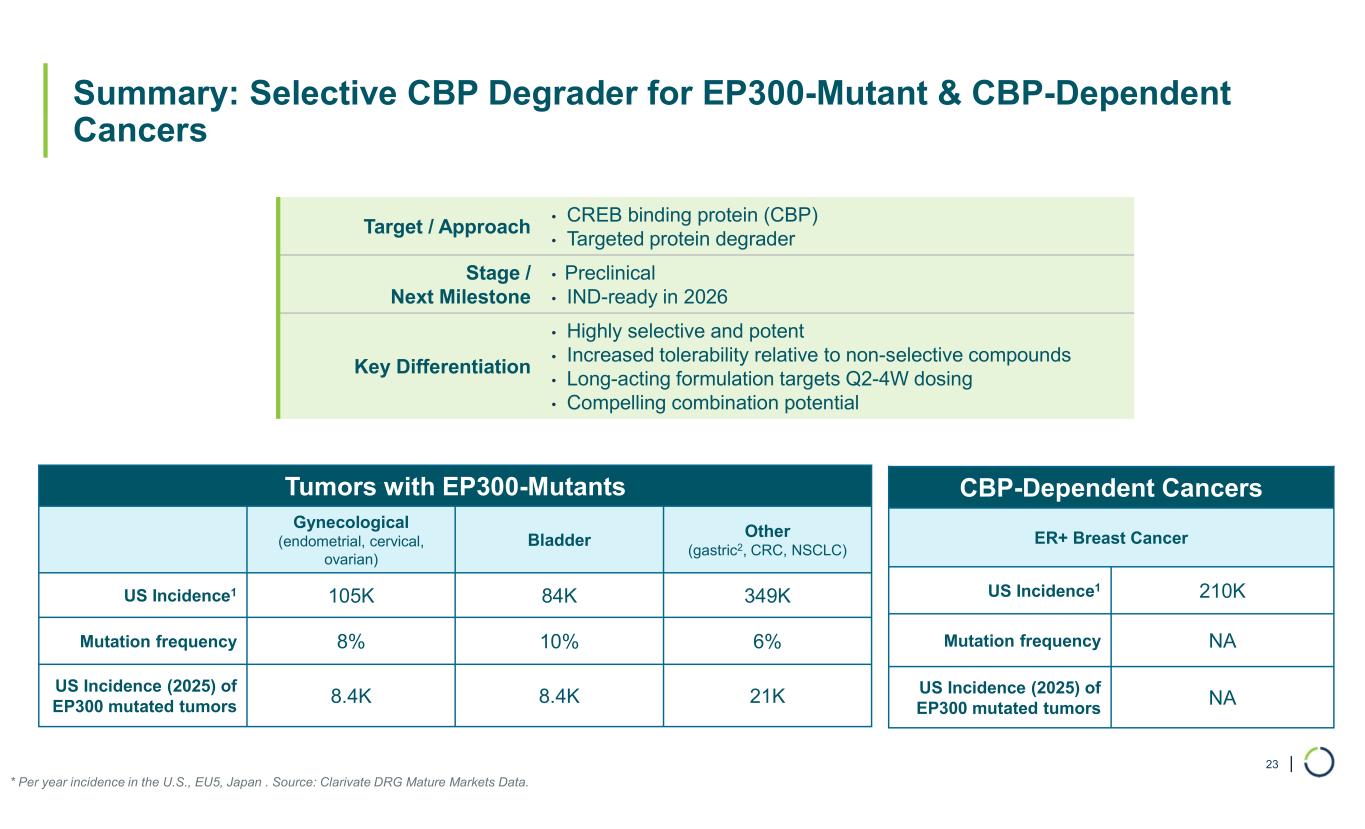
| Summary: Selective CBP Degrader for EP300-Mutant & CBP-Dependent Cancers 23 * Per year incidence in the U.S., EU5, Japan . Source: Clarivate DRG Mature Markets Data. • CREB binding protein (CBP) • Targeted protein degrader Target / Approach • Preclinical • IND-ready in 2026 Stage / Next Milestone • Highly selective and potent • Increased tolerability relative to non-selective compounds • Long-acting formulation targets Q2-4W dosing • Compelling combination potential Key Differentiation Tumors with EP300-Mutants Other (gastric2, CRC, NSCLC) Bladder Gynecological (endometrial, cervical, ovarian) 349K84K105KUS Incidence1 6%10%8%Mutation frequency 21K8.4K8.4K US Incidence (2025) of EP300 mutated tumors CBP-Dependent Cancers ER+ Breast Cancer 210KUS Incidence1 NAMutation frequency NA US Incidence (2025) of EP300 mutated tumors
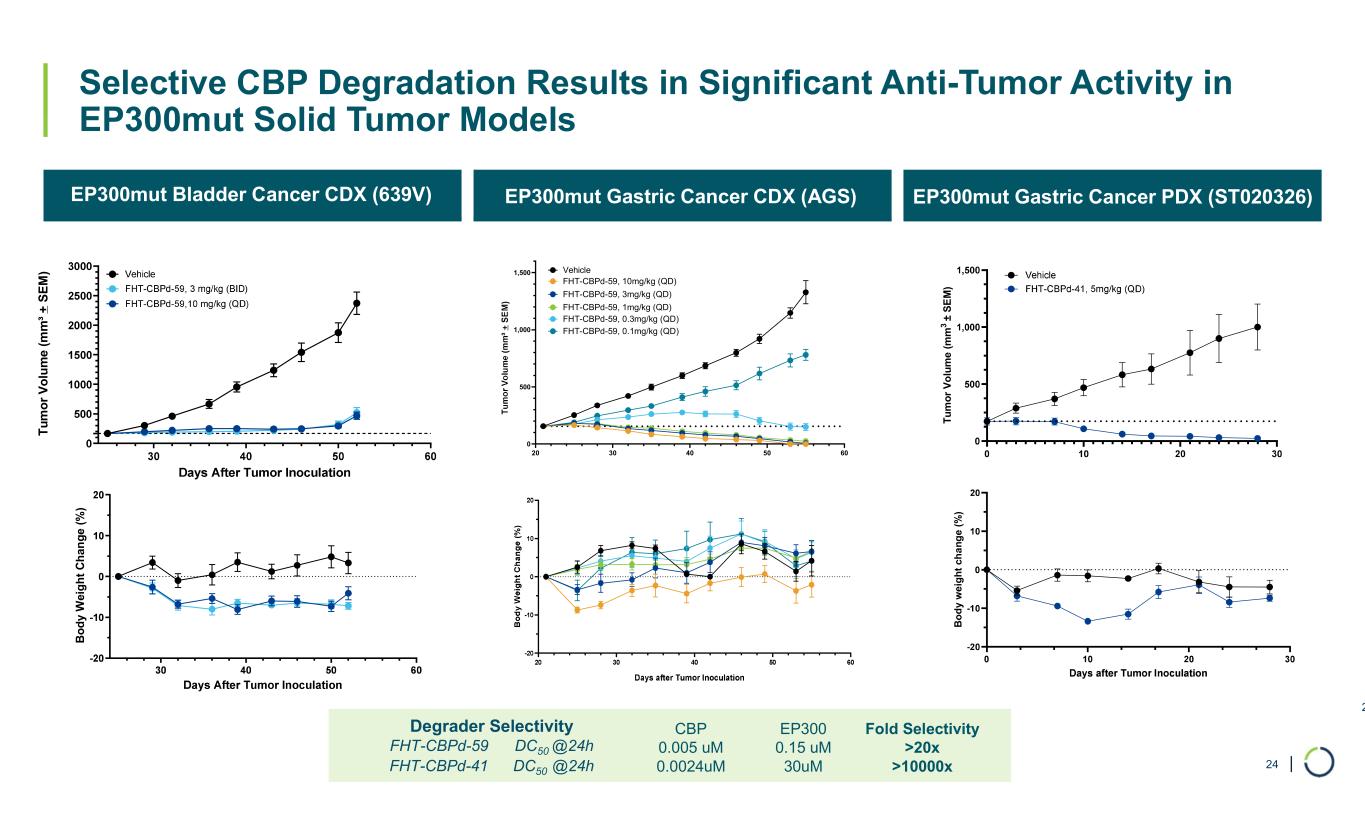
| Selective CBP Degradation Results in Significant Anti-Tumor Activity in EP300mut Solid Tumor Models 24 EP300mut Bladder Cancer CDX (639V) EP300mut Gastric Cancer CDX (AGS) EP300mut Gastric Cancer PDX (ST020326) T u m o r V o lu m e (m m ³ + S E M ) B o d y W ei g h t C h an g e ( % ) 20 30 40 50 60 0 500 1,000 1,500 Vehicle FHT-CBPd-59, 10mg/kg (QD) FHT-CBPd-59, 3mg/kg (QD) FHT-CBPd-59, 1mg/kg (QD) FHT-CBPd-59, 0.3mg/kg (QD) FHT-CBPd-59, 0.1mg/kg (QD) T u m o r V o lu m e ( m m 3 ± S E M ) B o d y w e ig h t ch a n g e ( % ) Fold Selectivity >20x >10000x EP300 0.15 uM 30uM CBP 0.005 uM 0.0024uM Degrader Selectivity FHT-CBPd-59 DC50 @24h FHT-CBPd-41 DC50 @24h 24
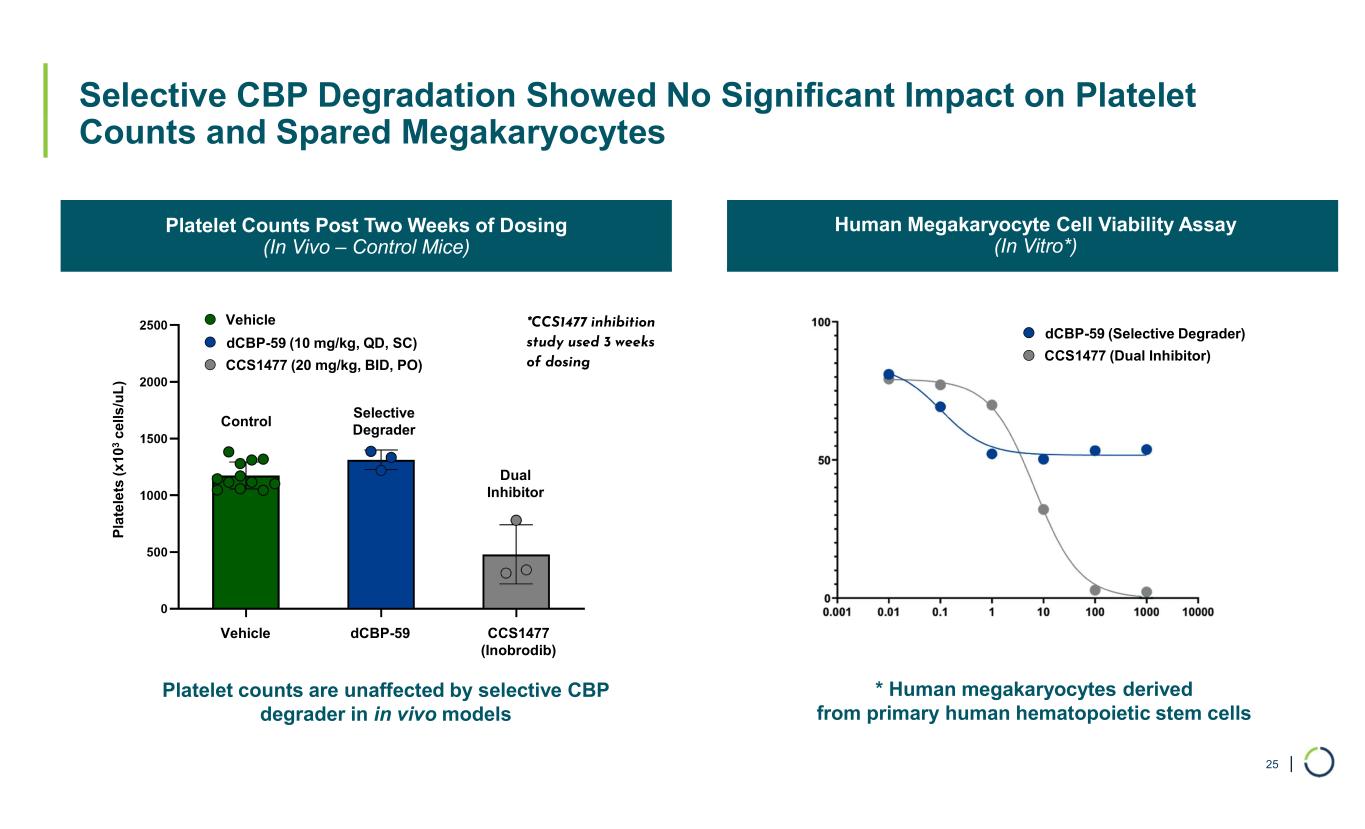
| Selective CBP Degradation Showed No Significant Impact on Platelet Counts and Spared Megakaryocytes 25 Platelet Counts Post Two Weeks of Dosing (In Vivo – Control Mice) Human Megakaryocyte Cell Viability Assay (In Vitro*) dCBP-59 CCS1477 (Inobrodib) Vehicle dCBP-59 (10 mg/kg, QD, SC) Vehicle CCS1477 (20 mg/kg, BID, PO) Dual Inhibitor Selective Degrader Control P la te le ts ( x 10 3 c el ls /u L ) *CCS1477 inhibition study used 3 weeks of dosing dCBP-59 (Selective Degrader) CCS1477 (Dual Inhibitor) Platelet counts are unaffected by selective CBP degrader in in vivo models * Human megakaryocytes derived from primary human hematopoietic stem cells
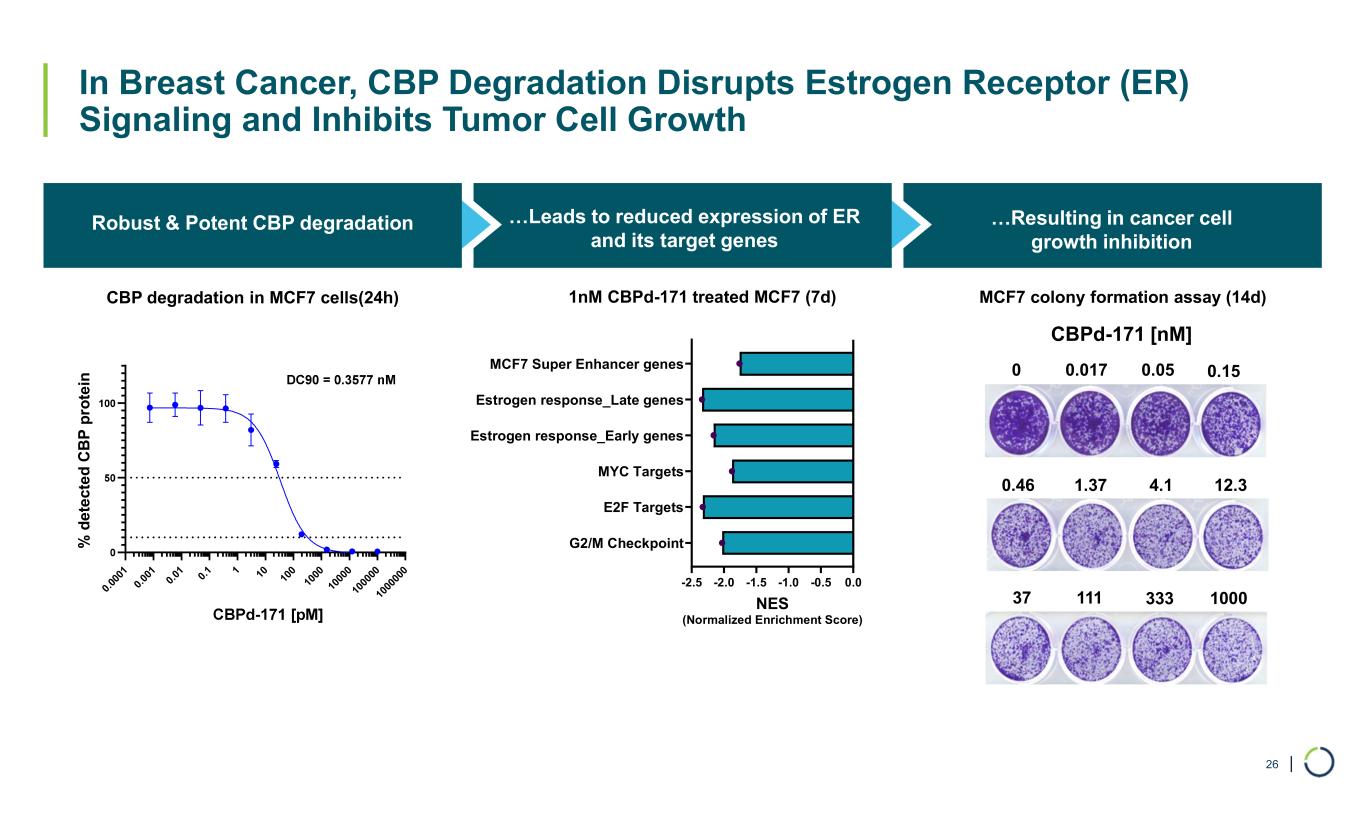
| In Breast Cancer, CBP Degradation Disrupts Estrogen Receptor (ER) Signaling and Inhibits Tumor Cell Growth CBP Robust & Potent CBP degradation …Resulting in cancer cell growth inhibition 26 0. 00 01 0. 00 1 0. 01 0. 1 1 10 10 0 10 00 10 00 0 10 00 00 10 00 00 0 % d e te ct e d C B P p ro te in CBP degradation in MCF7 cells(24h) …Leads to reduced expression of ER and its target genes MCF7 colony formation assay (14d) 33311137 12.34.11.370.46 0.150.050.017 1000 0 CBPd-171 [nM] -2.5 -2.0 -1.5 -1.0 -0.5 0.0 G2/M Checkpoint E2F Targets MYC Targets Estrogen response_Early genes Estrogen response_Late genes MCF7 Super Enhancer genes NES (Normalized Enrichment Score) 1nM CBPd-171 treated MCF7 (7d)
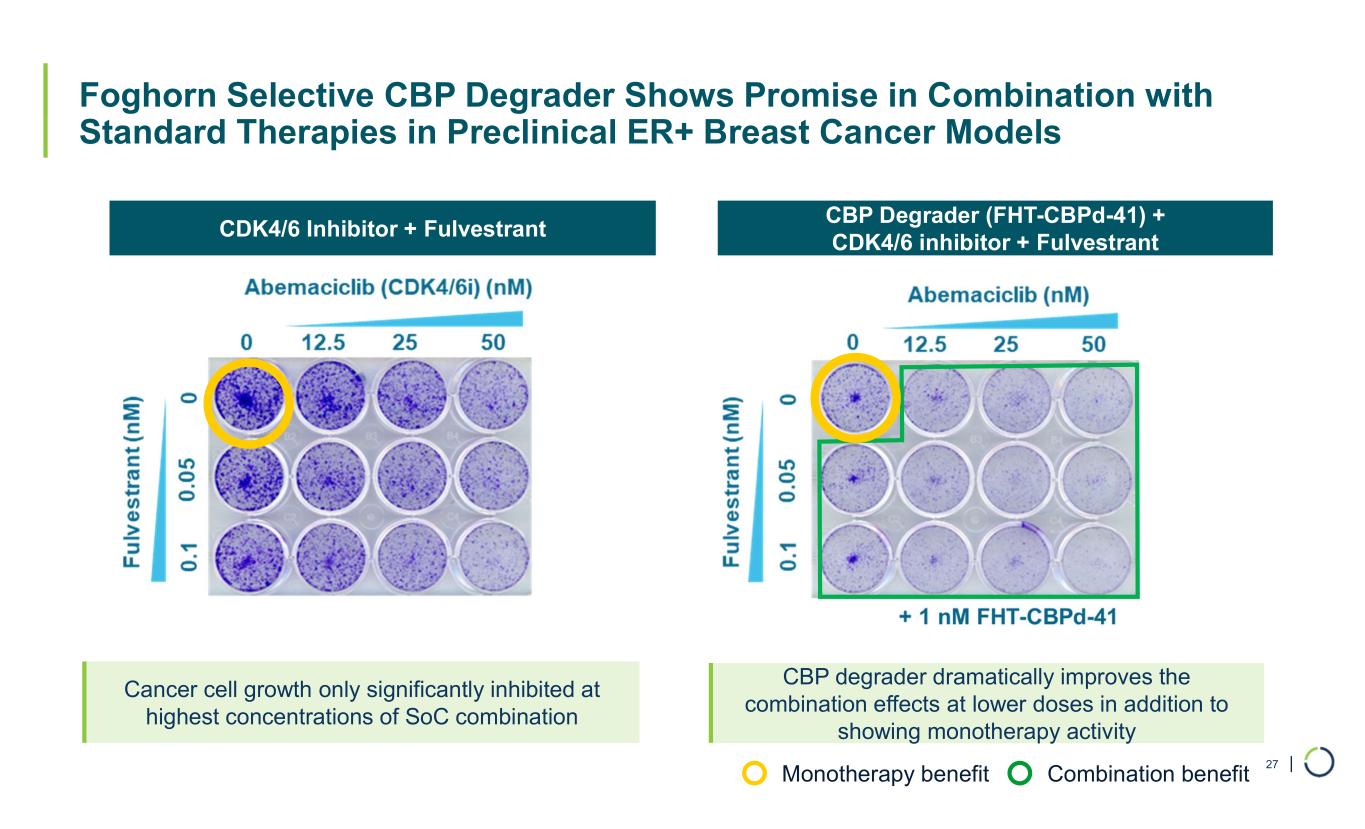
| Foghorn Selective CBP Degrader Shows Promise in Combination with Standard Therapies in Preclinical ER+ Breast Cancer Models 27 CDK4/6 Inhibitor + Fulvestrant CBP Degrader (FHT-CBPd-41) + CDK4/6 inhibitor + Fulvestrant Cancer cell growth only significantly inhibited at highest concentrations of SoC combination CBP degrader dramatically improves the combination effects at lower doses in addition to showing monotherapy activity Monotherapy benefit Combination benefit
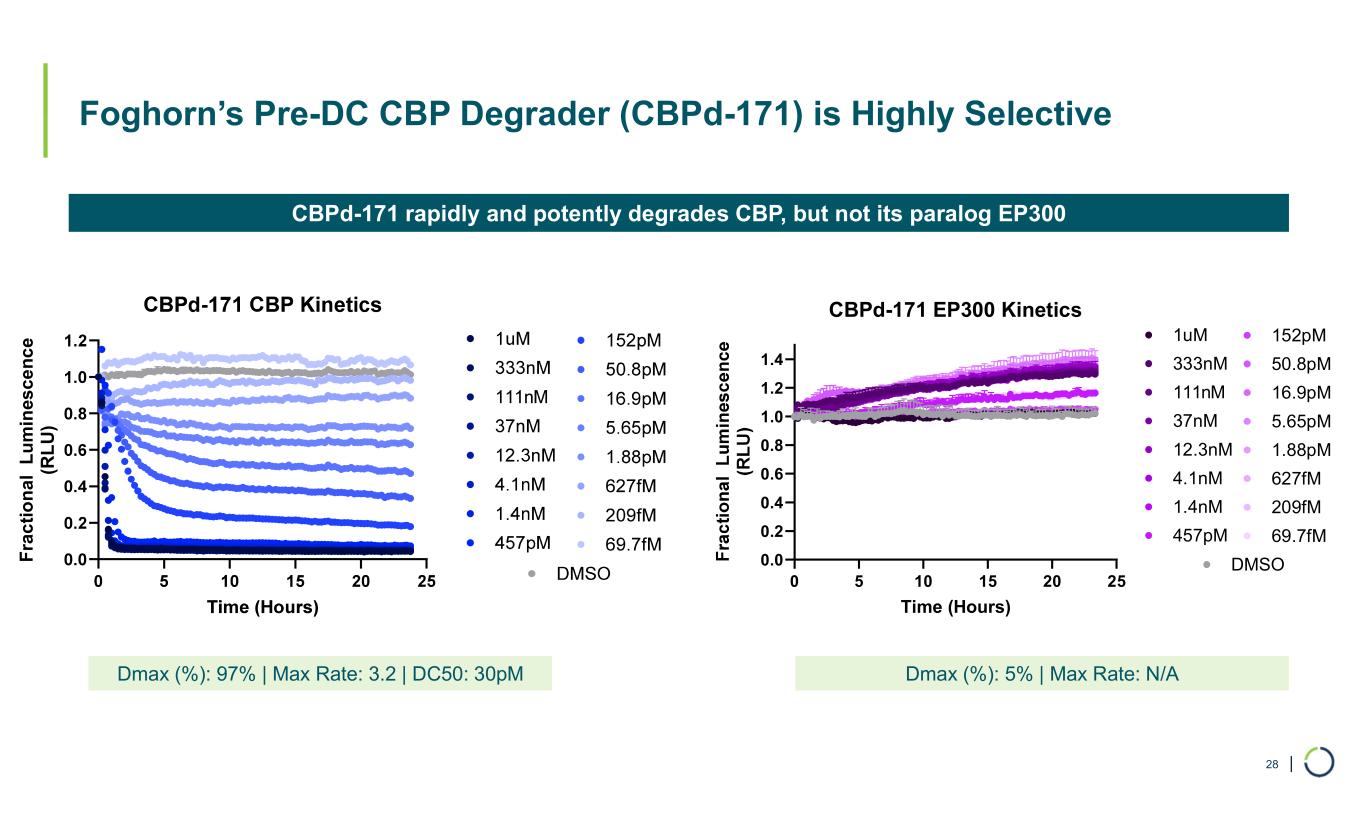
| Foghorn’s Pre-DC CBP Degrader (CBPd-171) is Highly Selective 28 CBPd-171 rapidly and potently degrades CBP, but not its paralog EP300 Dmax (%): 97% | Max Rate: 3.2 | DC50: 30pM Dmax (%): 5% | Max Rate: N/A F ra c ti o n a l L u m in es c e n c e (R L U ) 0 5 10 15 20 25 0.0 0.2 0.4 0.6 0.8 1.0 1.2 1.4 CBPd-171 EP300 Kinetics Time (Hours) DMSO 1uM 333nM 111nM 37nM 12.3nM 4.1nM 1.4nM 457pM 152pM 50.8pM 16.9pM 5.65pM 1.88pM 627fM 209fM 69.7fM
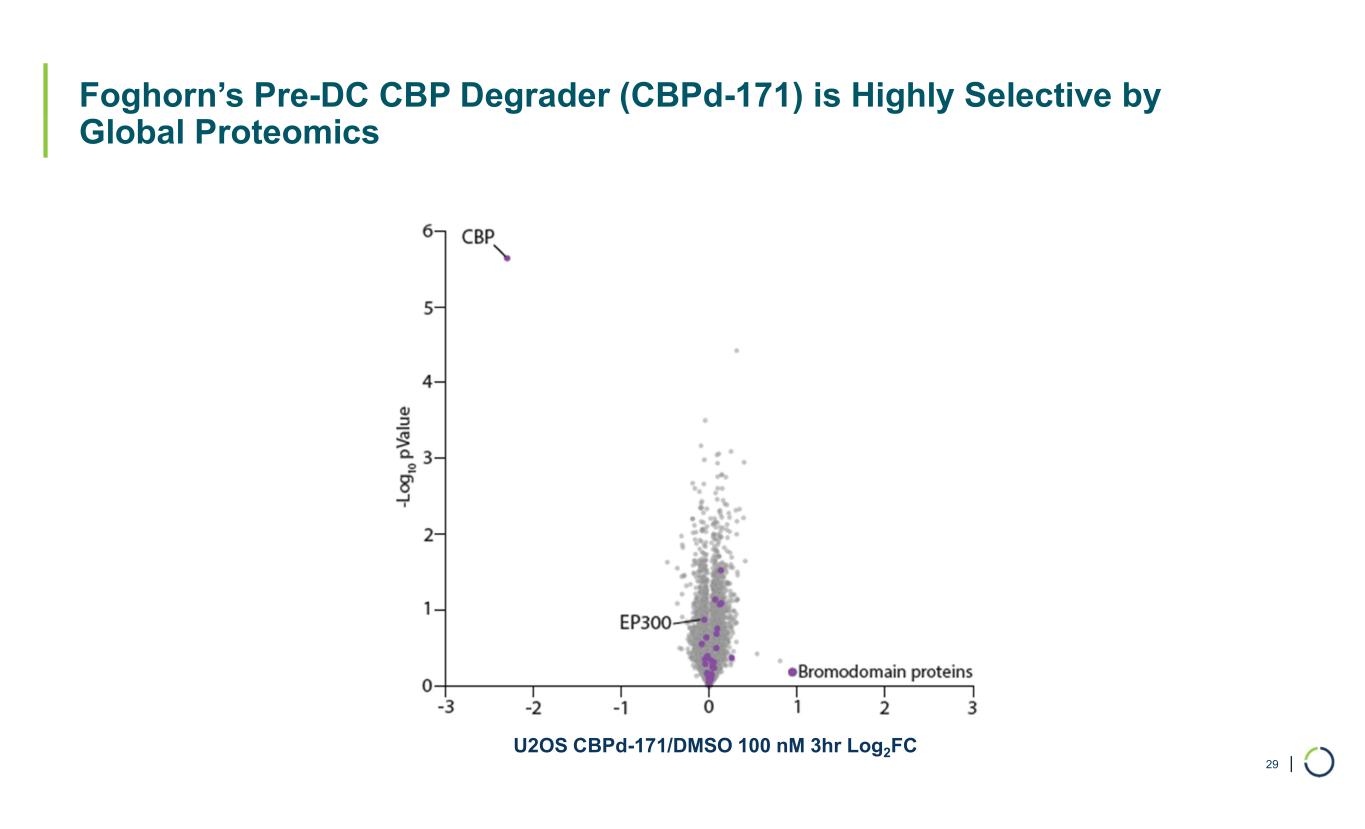
| Foghorn’s Pre-DC CBP Degrader (CBPd-171) is Highly Selective by Global Proteomics 29 U2OS CBPd-171/DMSO 100 nM 3hr Log2FC
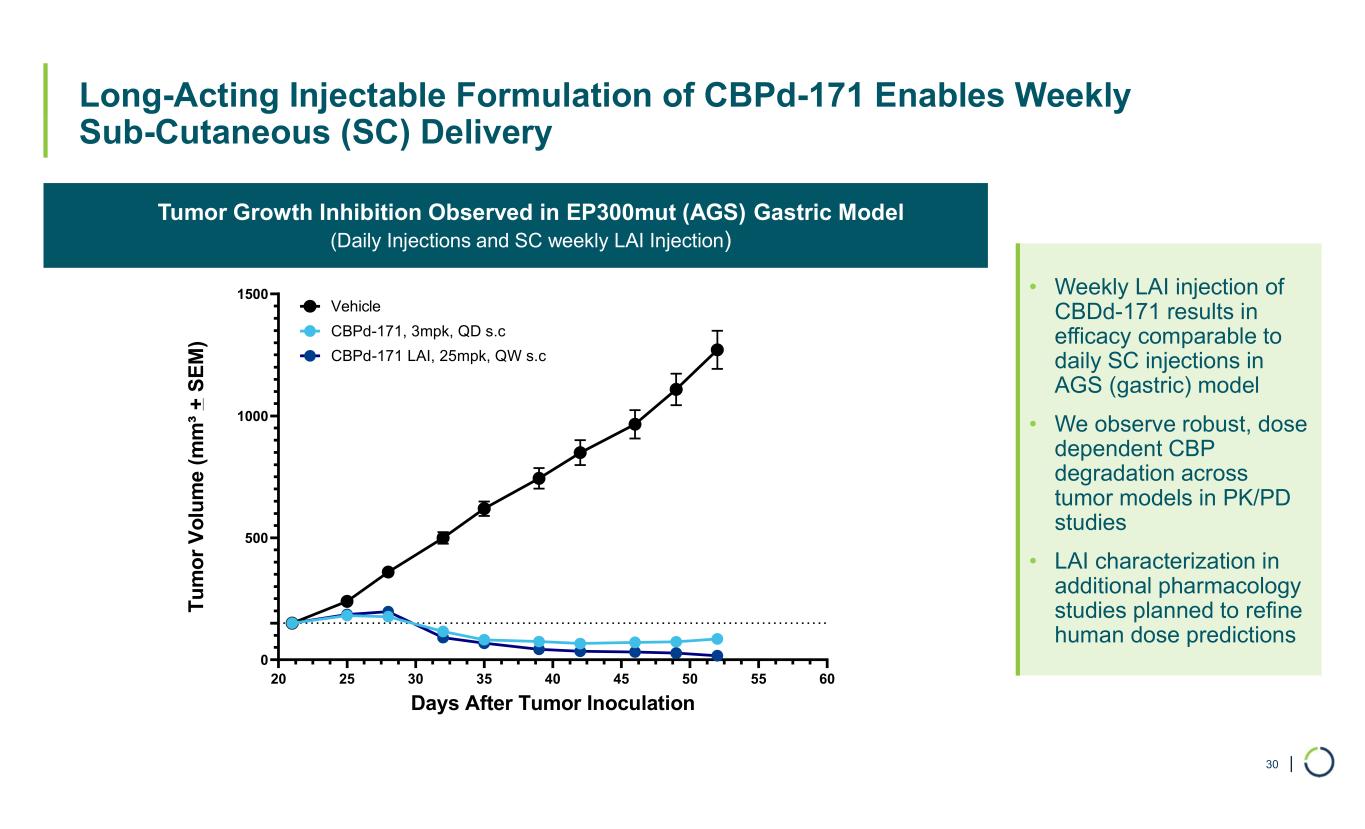
| Long-Acting Injectable Formulation of CBPd-171 Enables Weekly Sub-Cutaneous (SC) Delivery 30 Tumor Growth Inhibition Observed in EP300mut (AGS) Gastric Model (Daily Injections and SC weekly LAI Injection) • Weekly LAI injection of CBDd-171 results in efficacy comparable to daily SC injections in AGS (gastric) model • We observe robust, dose dependent CBP degradation across tumor models in PK/PD studies • LAI characterization in additional pharmacology studies planned to refine human dose predictions 20 25 30 35 40 45 50 55 60 0 500 1000 1500 Days After Tumor Inoculation T u m o r V o lu m e ( m m ³ + S E M ) CBPd-171, 3mpk, QD s.c Vehicle CBPd-171 LAI, 25mpk, QW s.c

Selective EP300 Degrader For CBP-Mutant and EP300-Dependent Cancers 31

| Summary: Selective EP300 Degrader for CBP-Mutant & EP300-Dependent Cancers 32 * Per year incidence in the U.S., EU5, Japan. Source: Clarivate DRG Mature Markets Data. • E1A binding protein p300 (EP300) • Targeted protein degrader Target / Approach • Broad range of heme malignancies focused on Multiple Myeloma and DLBCL • AR+ prostate • Bladder, melanoma, others Indications • Preclinical • IND-enabling studies in 2026 Stage / Next Milestone • Deeper efficacy response with selective degrader vs non-selective molecules • Improved tolerability profile vs non-selective molecules • Patient selection biomarker for Diffuse Large B-Cell Lymphoma (DLBCL) Key Differentiation EP300 Dependency in Hematological Malignancies AML + MDSDLBCLMultiple Myeloma 37K32K31KUS Incidence1
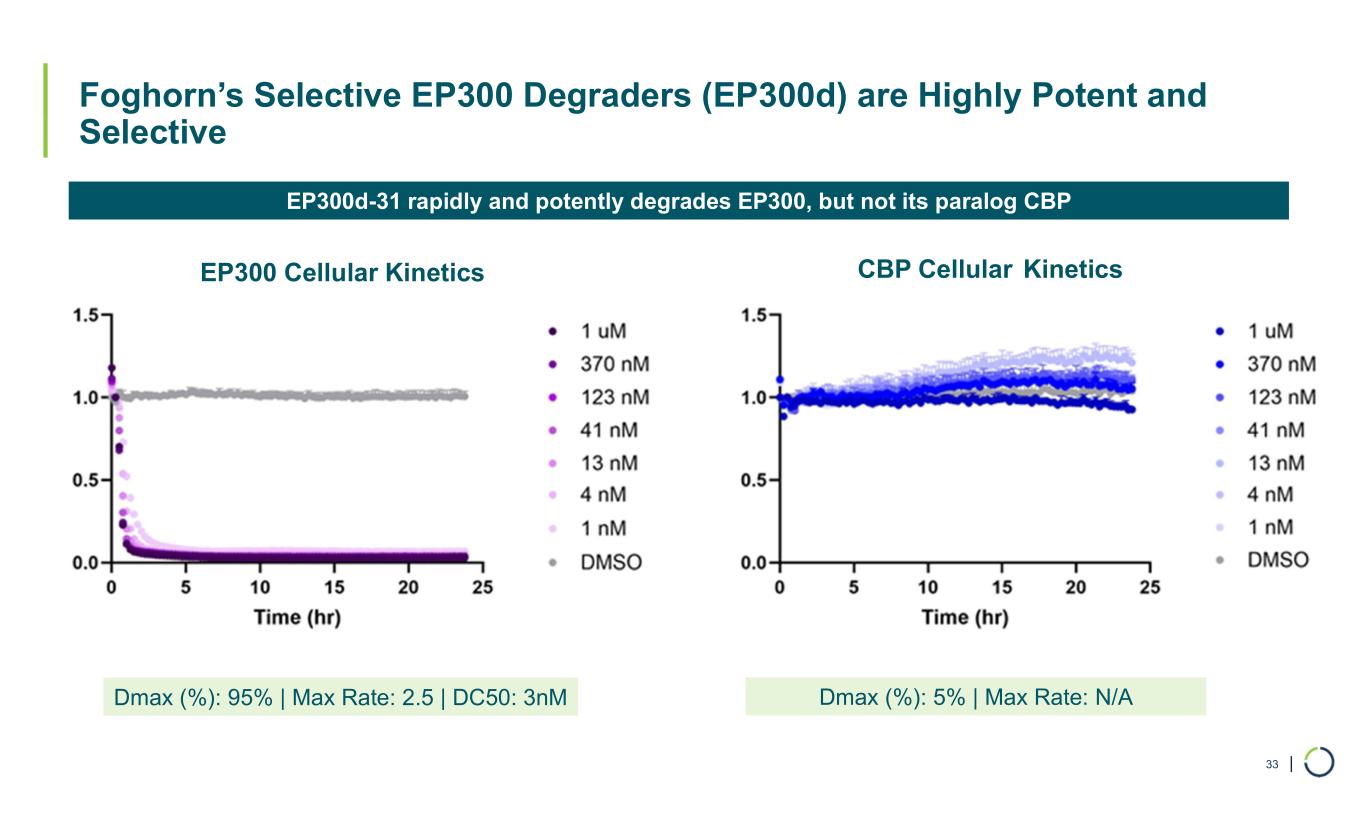
| Foghorn’s Selective EP300 Degraders (EP300d) are Highly Potent and Selective 33 Dmax (%): 95% | Max Rate: 2.5 | DC50: 3nM Dmax (%): 5% | Max Rate: N/A EP300 Cellular Kinetics CBP Cellular Kinetics EP300d-31 rapidly and potently degrades EP300, but not its paralog CBP

| Foghorn’s EP300 Degrader (EP300d-79) is Highly Selective by Global Proteomics 34 U2OS EP300d/DMSO 300 nM 6hr Log2FC

| EP300 Degradation Shows Anti-Proliferative Activity in Broad Range of Hematological Malignancies 35MM: Multiple Myeloma; DLBCL: Diffuse Large B-Cell Lymphoma; AML: Acute Myeloid Leukemia; BL: Burkitt’s Lymphoma; TCL: T-cell Lymphomas; T-ALL: T-cell Acute Lymphoblastic Leukemia; B-ALL: B-cell Acute Lymphoblastic Leukemia; CML: Chronic Myeloid Leukemia; HL: Hodgkin Lymphoma; MCL: Mantle Cell Lymphoma Anti-Tumor Activity Across Full Range of Heme Sub-Lineages (~ 70% of All Tested Cell Lines are Sensitive)
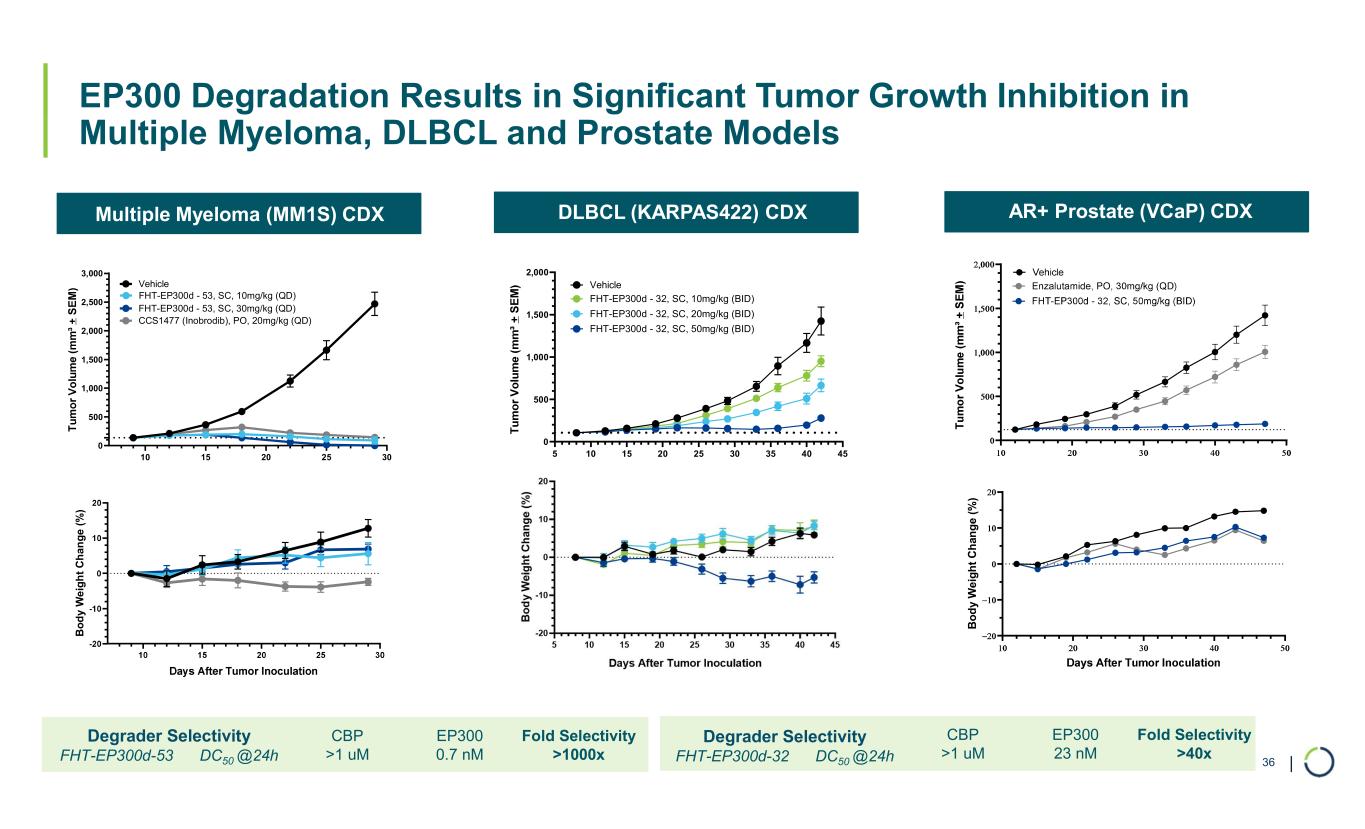
| Multiple Myeloma (MM1S) CDX DLBCL (KARPAS422) CDX AR+ Prostate (VCaP) CDX EP300 Degradation Results in Significant Tumor Growth Inhibition in Multiple Myeloma, DLBCL and Prostate Models 36 B o d y W ei g h t C h an g e (% ) 5 10 15 20 25 30 35 40 45 0 500 1,000 1,500 2,000 Vehicle FHT-EP300d - 32, SC, 50mg/kg (BID) Days After Tumor Inoculation FHT-EP300d - 32, SC, 20mg/kg (BID) FHT-EP300d - 32, SC, 10mg/kg (BID) B o d y W e ig h t C h an g e ( % ) 10 15 20 25 30 0 500 1,000 1,500 2,000 2,500 3,000 Days After Tumor Inoculation Vehicle FHT-EP300d - 53, SC, 30mg/kg (QD) FHT-EP300d - 53, SC, 10mg/kg (QD) CCS1477 (Inobrodib), PO, 20mg/kg (QD) Fold Selectivity >1000x EP300 0.7 nM CBP >1 uM Degrader Selectivity FHT-EP300d-53 DC50 @24h Fold Selectivity >40x EP300 23 nM CBP >1 uM Degrader Selectivity FHT-EP300d-32 DC50 @24h

| EP300 Degradation Modulates the Transcription Factors That Multiple Myeloma Depends On 37 EP300 degradation downregulates the expression of several transcription factors H929 OPM2 Lineage-dependence on EP300-regulated transcription factors KMS11RPMI8226 OPM2 RPMI8226 KMS11 dEP300-9 treatment, cell line-specific DC90+ doses (20-300nM), 6h

| Selective EP300 Degradation Induces Apoptosis in MM 38 Treatment with a selective EP300 degrader (dEP300-079) results in time- and concentration- dependent induction of apoptosis MM1S cell line
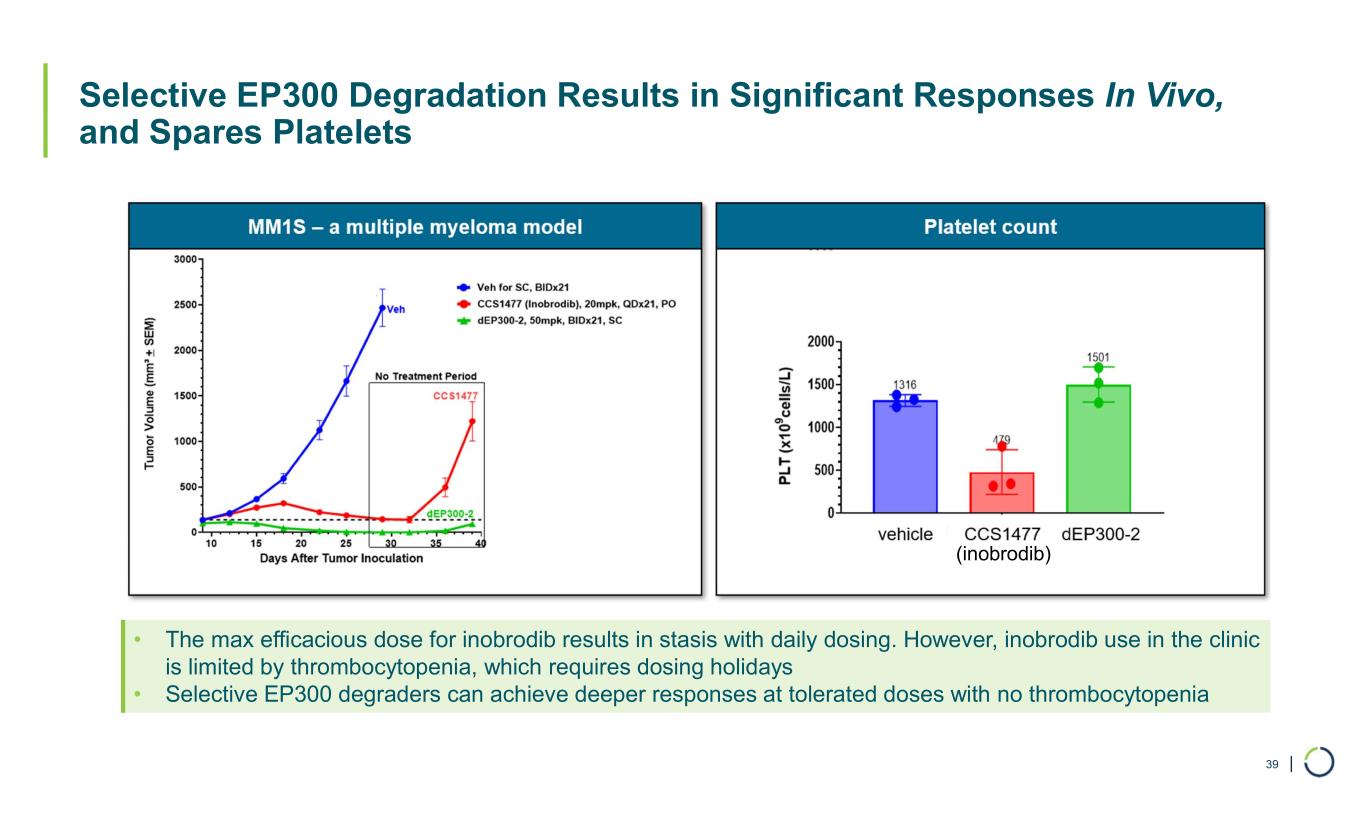
| Selective EP300 Degradation Results in Significant Responses In Vivo, and Spares Platelets 39 • The max efficacious dose for inobrodib results in stasis with daily dosing. However, inobrodib use in the clinic is limited by thrombocytopenia, which requires dosing holidays • Selective EP300 degraders can achieve deeper responses at tolerated doses with no thrombocytopenia (inobrodib)

| VHL-Based Selective EP300 Degrader Maintains Activity in IMiD Resistant Cell Lines 40 Resistant MM1S cell lines were developed through 5–6 months of in vitro exposure to gradually increasing concentrations of lenalidomide, pomalidomide, iberdomide, or mezigdomide dEP300-9 dEP300-9 pomalidomide lenalidomide mezigdomide iberdomide
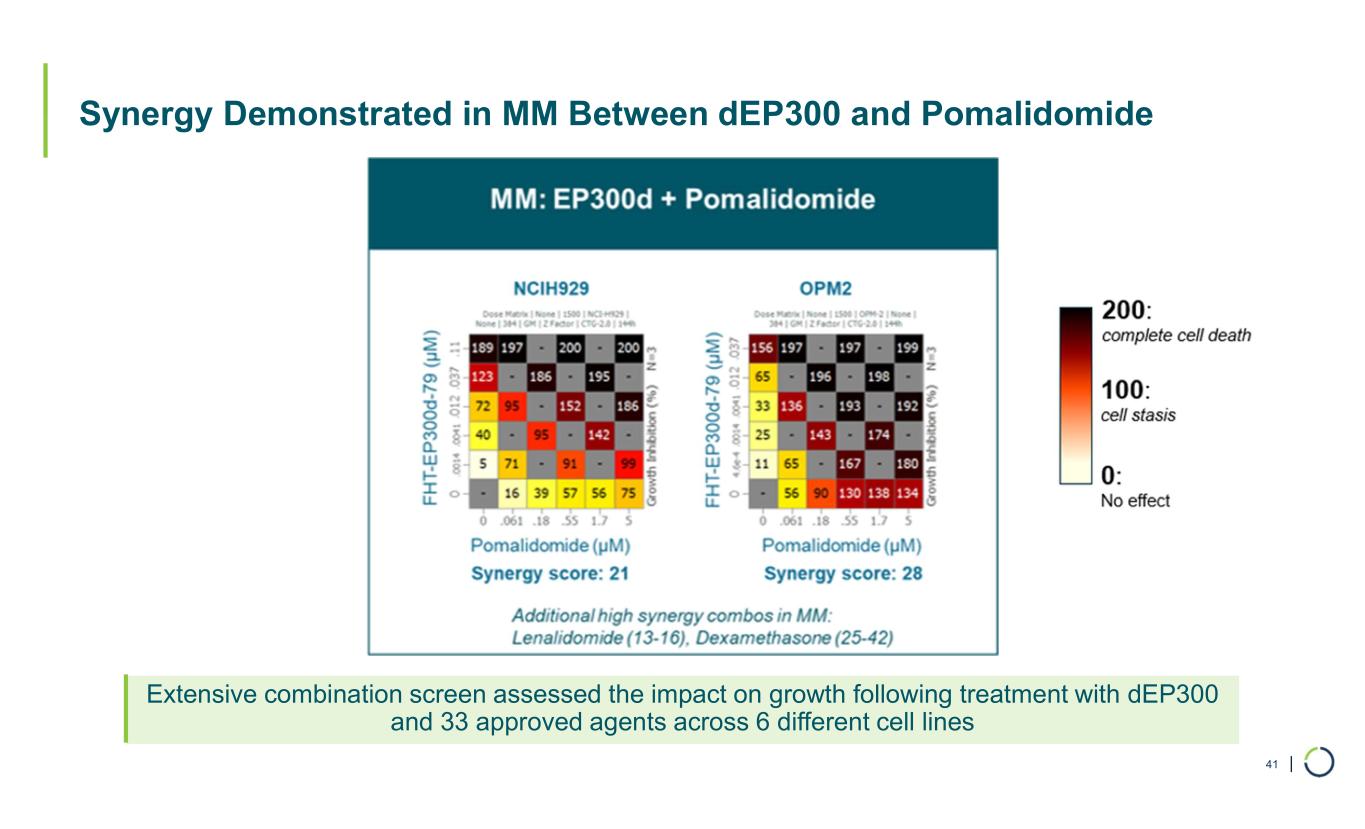
| Synergy Demonstrated in MM Between dEP300 and Pomalidomide 41 Extensive combination screen assessed the impact on growth following treatment with dEP300 and 33 approved agents across 6 different cell lines

| Selective EP300 Degraders Approaching Pre-DC CBACompound 0.3 [104%]0.3 [99%]0.4 [95%]U2OS EP300 uDC50 (nM) [Dmax%] On- Target <0.15Actioned2.1MM1S EP300 uDC90 (24 hr, nM) 0.152.00.61MM1S (nM, uGI50) 31 / >10,000102 / >10,0004 / 3000CBP / BRD4 TR-FRET IC50 (nM) Off-Target 356x [63%]17497x [--]26x [44%]U2OS CBP/EP DC50 [CBP Dmax%] 3,665x319x125xSUDHL/MM1s GI50 >30>30>30BRD4 DC50 (µM) [Dmax%] 244244296Kin Solubility (PBS, pH=7.4, µM) ADME 0 / 21 / 11<4 / 10 / 40 / 0 / 4LMS (m / r / h) (Clint, µL/min/mg) 1.6 / 2.6 8.0 / 3.8 0.73 / 0.26 Actioned 6.4 / 12.4 1.3 / 3.8 0.27 / 0.69 IV CL (mL/min/Kg) T1/2 (h) mouse /rat Vdss (L / Kg) 42

Selective ARID1B Degrader For ARID1A-Mutant Cancers 43

| ARID1B is a Major Synthetic Lethal Target with Potential in Up To 5% of All Solid Tumors 44 * Per year incidence in the U.S., EU5, Japan. Source: Clarivate DRG Mature Markets Data. • ARID1B • Targeted protein degrader Target / Approach • Preclinical • In vivo proof of concept in 2026 Stage / Next Milestone • Multiple ARID1B binders with nM affinity and selectivity • Selective ARID1B degradation Key Differentiation Tumors with ARID1A-Mutants NSCLCBladderGastric2 & GEJEndometrial 195K84K37K66KUS Incidence1 7%24%20%38%Mutation frequency 14K20K7K25K US Incidence (2025) of ARID1A-mutant tumors

| ARID1B: Drugging A Previously Undruggable Target • Large and highly unstructured protein ~ 240 kDa • No known enzymatic function • Member of large, multi-subunit complex • High sequence homology (~60%) to ARID1A 45 ARID1A/B BAF Nucleosome PDB ID: 6LTJ • Discover binders to ARID1B • Use binders to develop bifunctional degraders Drug Targeting Considerations Approach
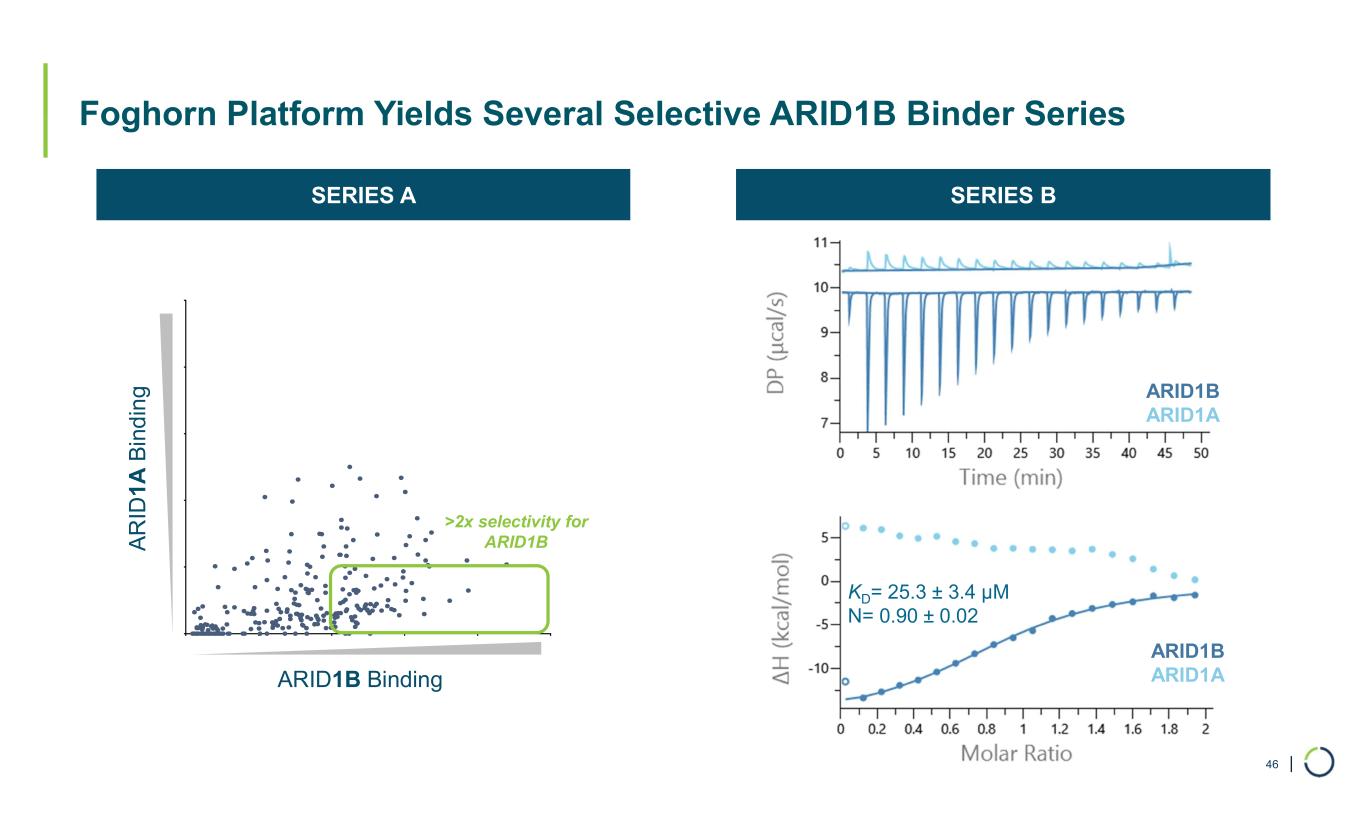
| Foghorn Platform Yields Several Selective ARID1B Binder Series 46 ARID1B ARID1A KD= 25.3 ± 3.4 µM N= 0.90 ± 0.02 ARID1B ARID1A >2x selectivity for ARID1B ARID1B Binding A R ID 1 A B in d in g SERIES A SERIES B
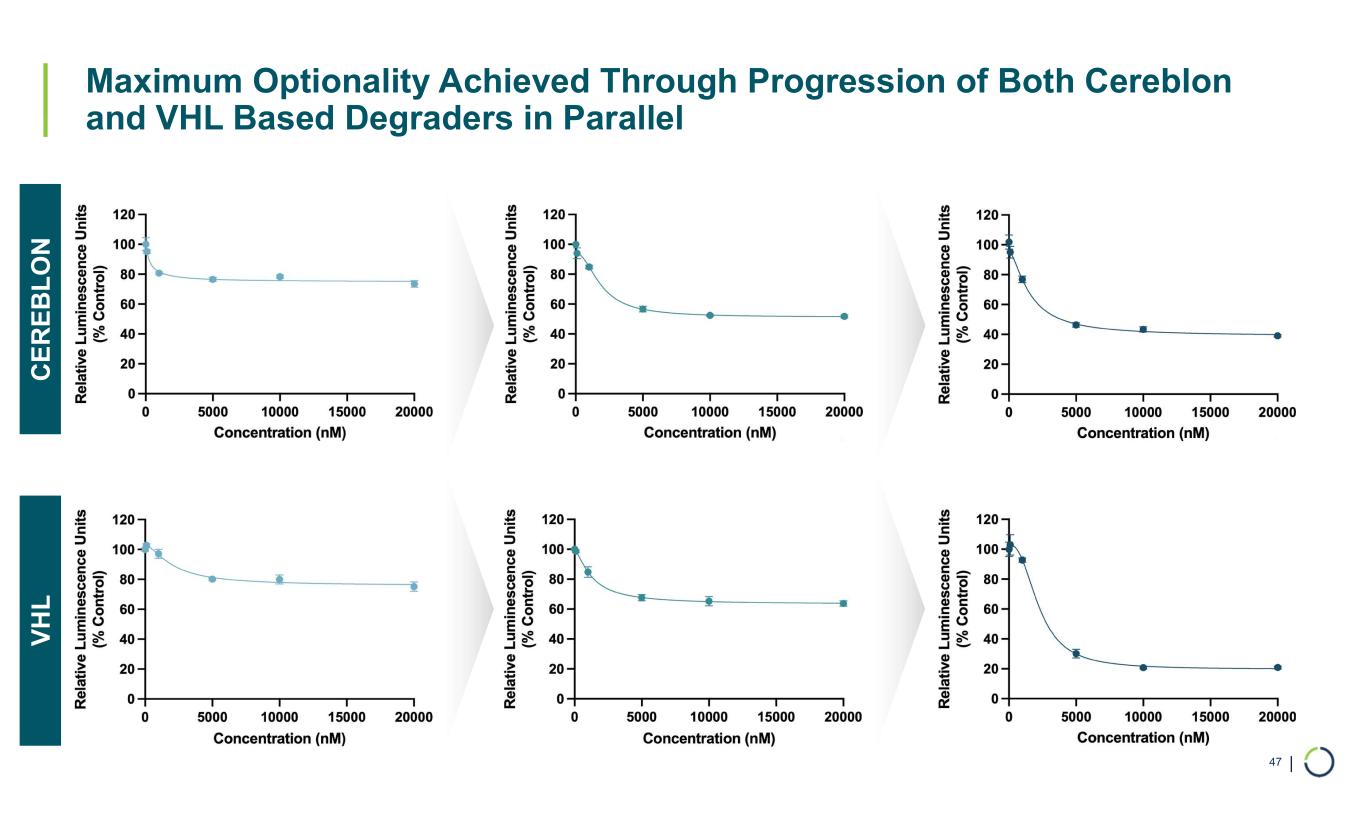
| Maximum Optionality Achieved Through Progression of Both Cereblon and VHL Based Degraders in Parallel 47 C E R E B L O N V H L

| Lead ARID1B Degrader Demonstrates Robust Degradation Across Multiple Cell Lines 48 IMMUNOBLOT HiBiT - ENDPOINT HCT116 (Colorectal)HT-1376 (Bladder) HiBiT – KINETICS HCT116 (Colorectal) 24 hours 24 hours R el a ti v e L u m in e sc e n c e U n it s (% C o n tr o l)
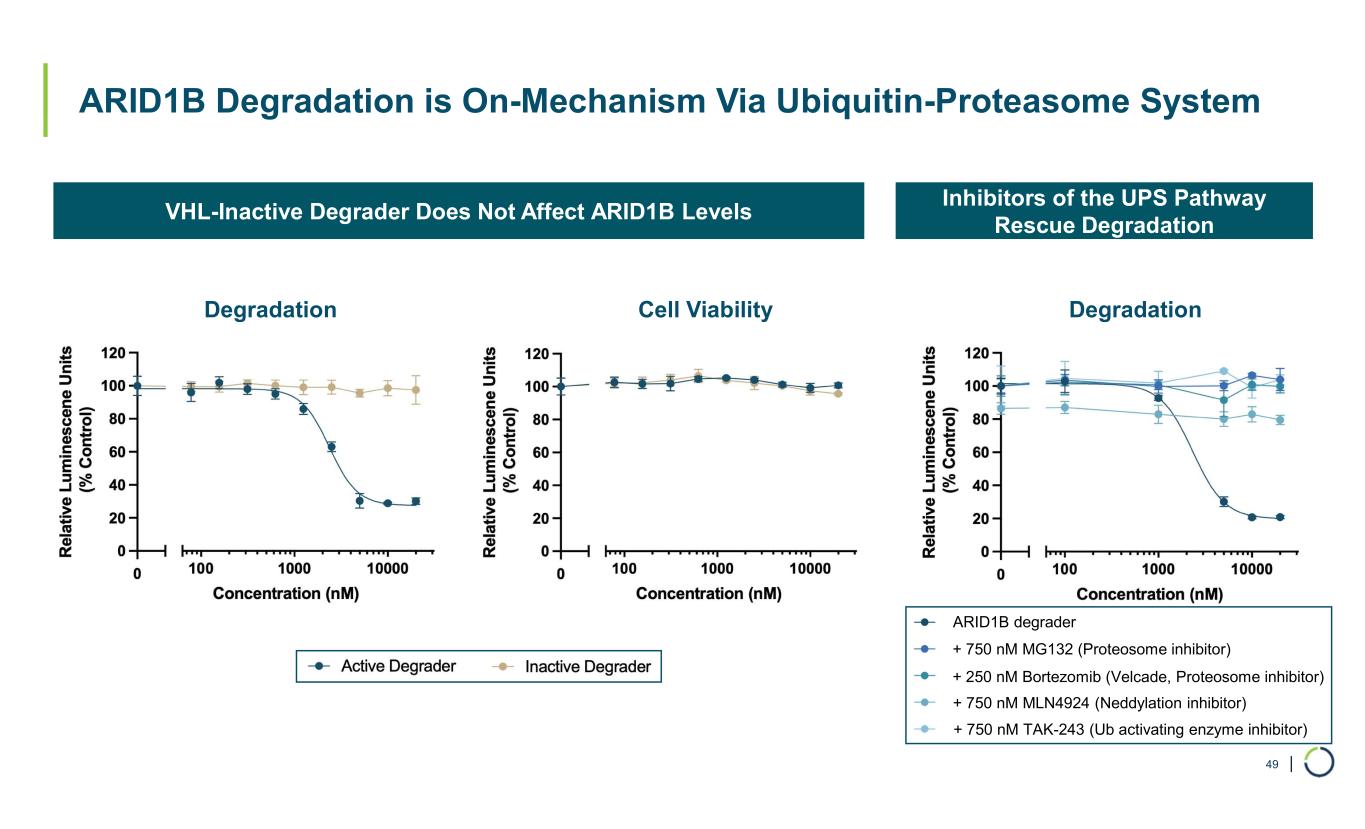
| ARID1B Degradation is On-Mechanism Via Ubiquitin-Proteasome System 49 Inhibitors of the UPS Pathway Rescue Degradation VHL-Inactive Degrader Does Not Affect ARID1B Levels Degradation Cell Viability Degradation + 750 nM MG132 (Proteosome inhibitor) + 250 nM Bortezomib (Velcade, Proteosome inhibitor) + 750 nM MLN4924 (Neddylation inhibitor) + 750 nM TAK-243 (Ub activating enzyme inhibitor) ARID1B degrader
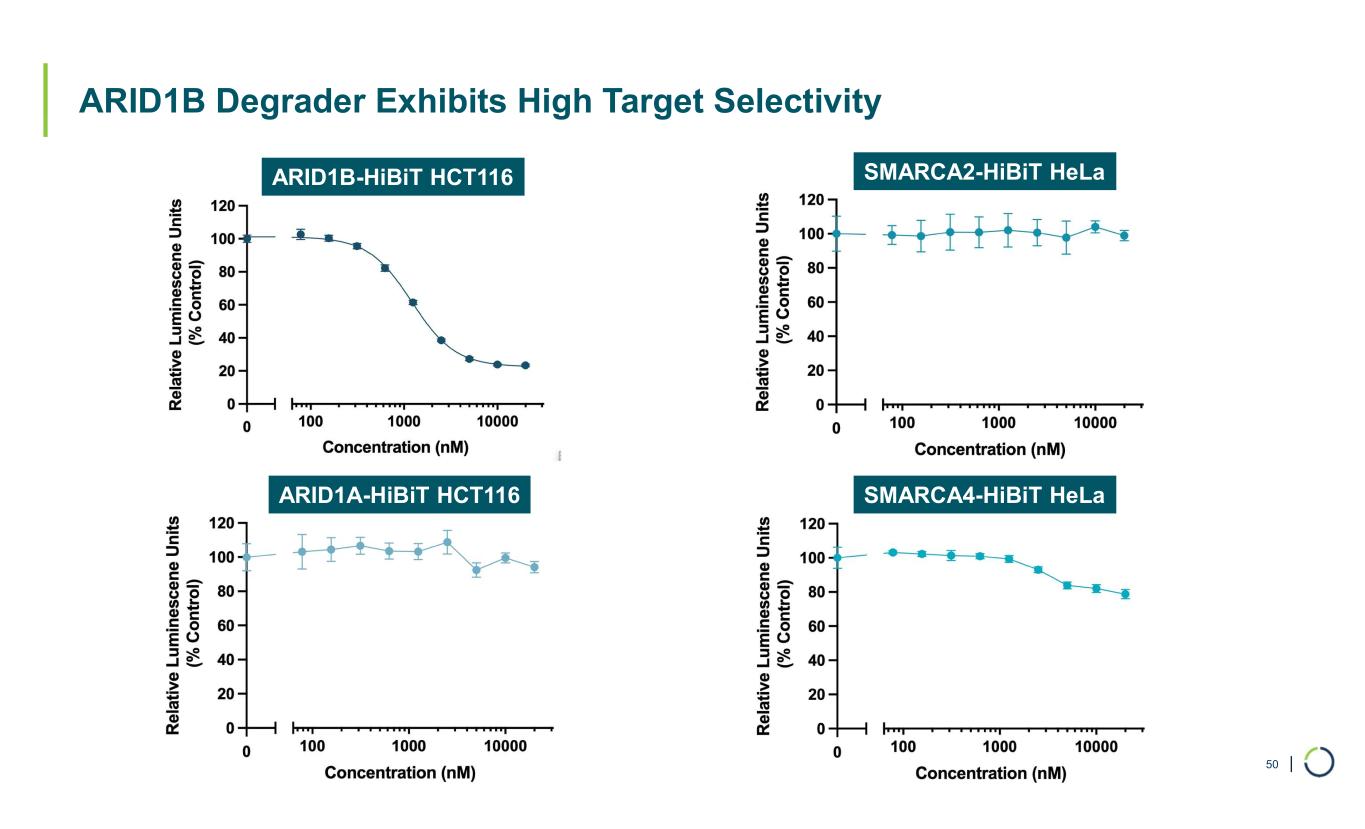
| ARID1B Degrader Exhibits High Target Selectivity 50 ARID1B-HiBiT HCT116 ARID1A-HiBiT HCT116 SMARCA2-HiBiT HeLa SMARCA4-HiBiT HeLa

| Selective ARID1B Degradation Demonstrated by Global Proteomics 51

| 0.0 0.2 0.4 0.6 0.8 1.0 1 2 3 4 5 6 Selective ARID1B Degradation has Demonstrated Effects on Downstream Target Genes – Progressing Towards Proof of Concept 52 3 Day ARID1B shRNA Treatment ARID1B Degrader Treatment y = 0.6782x + 0.2591 R² = 0.7754 0.00 0.20 0.40 0.60 0.80 1.00 1.20 0.0 0.5 1.0 ARID1B expression y = -2.6042x + 3.1832 R² = 0.8159 0.0 0.5 1.0 1.5 2.0 2.5 3.0 3.5 0.0 0.5 1.0 ARID1B expression ARID1B Gene Expression ARID1B Expression ARID1B ExpressionRange of Dox Treatment R e la ti v e E xp re ss io n R e la ti v e E xp re ss io n R e la ti v e E xp re ss io n Down Gene A Expression Up Gene B Expression 24h Treatment 48h Treatment Down Gene A Expression Up Gene B Expression Putative target genes identified from literature and confirmed via internal shRNA efforts

| Advancing Multiple Programs with Near-Term Milestones 53 Phase 1 Monotherapy DataFHD-909* (LY4050784) (Selective SMARCA2 Inhibitor) 2026IND Ready Selective CBP Degrader 2026Development CandidateSelective EP300 Degrader ConfidentialTransition to Lilly / INDSelective SMARCA2 Degrader* ConfidentialTransition to Lilly / INDLilly Target #2** 2026In Vivo Proof of ConceptSelective ARID1B Degrader * 50/50 U.S. economic split, ex-U.S. royalties. ** Pending Lilly decision to proceed, 50/50 U.S. economic split, ex-U.S. royalties. Confidential

| Developing First-in-Class Precision Medicines Targeting Major Unmet Needs in Cancer 54 Large Market Potential Chromatin biology is implicated in up to 50% of tumors, potentially impacting ~2.5 million patients Foghorn’s current pipeline potentially addresses more than 500,000 of these patients Broad pipeline across a range of targets and small molecule modalities Major Strategic Collaboration Strategic collaboration with Lilly; $380 million upfront; 50/50 U.S. economic split on two lead programs Well- Funded $180.3 million in cash and equivalents (as of 09/30/2025) Cash runway into 2028 Shares outstanding: approximately 63.0M* Value Drivers Selective SMARCA2 Inhibitor, FHD-909, partnered with Lilly, in Phase 1 trial Advancement of preclinical assets (Selective SMARCA2, CBP, EP300, ARID1B Degraders) towards INDs Protein degrader platform with expansion into induced proximity Leader in Unique Area of Cancer Biology Foghorn is a leader in targeting chromatin biology, which has the potential to address underlying dependencies of many genetically defined cancers Platform with initial focus in oncology, therapeutic area expansion potential *Includes common shares outstanding and pre-funded warrants as of 09/30/2025.

Unique biology Precision therapeutics Broad impact 55 November 2025

Appendix 56

SMARCA2 Program FHD-609 is a Selective, Potent, Protein Degrader of the BRD9 component of the BAF complex 57
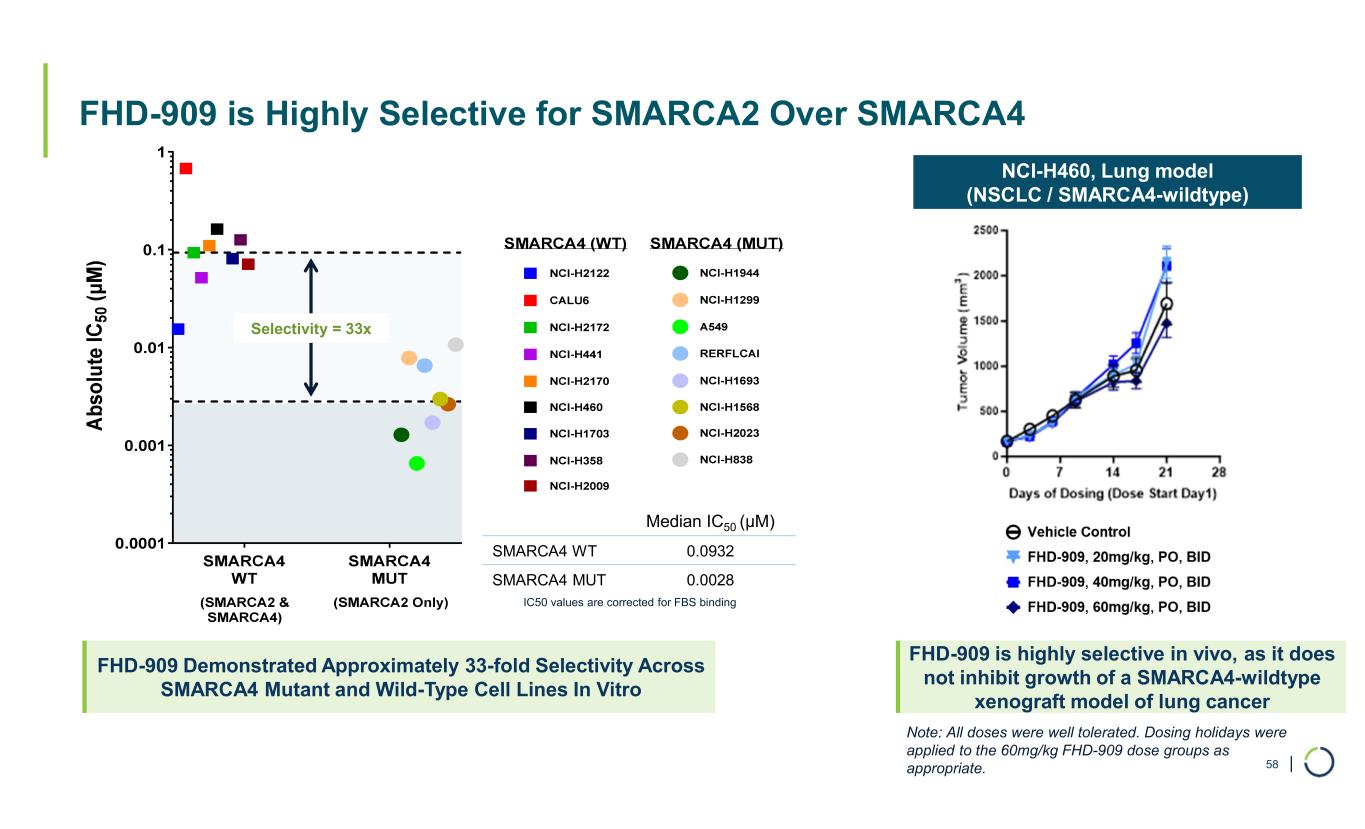
| FHD-909 is Highly Selective for SMARCA2 Over SMARCA4 58 FHD-909 Demonstrated Approximately 33-fold Selectivity Across SMARCA4 Mutant and Wild-Type Cell Lines In Vitro Selectivity = 33x Median IC50 (µM) 0.0932SMARCA4 WT 0.0028SMARCA4 MUT NCI-H460, Lung model (NSCLC / SMARCA4-wildtype) FHD-909 is highly selective in vivo, as it does not inhibit growth of a SMARCA4-wildtype xenograft model of lung cancer IC50 values are corrected for FBS binding Note: All doses were well tolerated. Dosing holidays were applied to the 60mg/kg FHD-909 dose groups as appropriate.
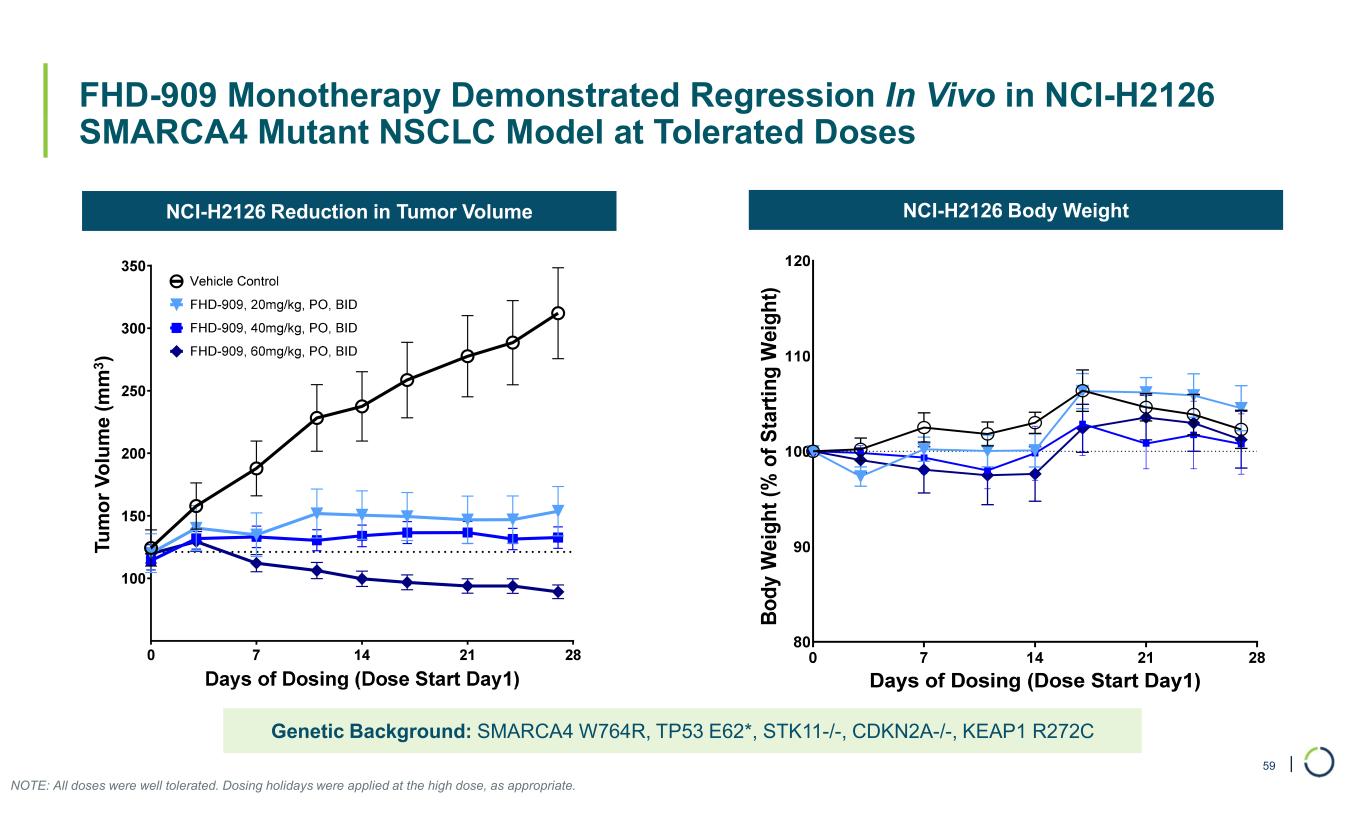
| FHD-909 Monotherapy Demonstrated Regression In Vivo in NCI-H2126 SMARCA4 Mutant NSCLC Model at Tolerated Doses 59 NCI-H2126 Reduction in Tumor Volume NCI-H2126 Body Weight Genetic Background: SMARCA4 W764R, TP53 E62*, STK11-/-, CDKN2A-/-, KEAP1 R272C NOTE: All doses were well tolerated. Dosing holidays were applied at the high dose, as appropriate. T u m o r vo lu m e (m m 3 ) Tu m o r V o lu m e (m m 3 )

| FHD-909 Monotherapy Demonstrated Strong In Vivo Activity Across SMARCA4 Mutant NSCLC Models at Tolerated Doses 60 0 7 14 21 28 100 200 300 400 500 Days of Dosing (Dose Start Day1) Vehicle Control FHD-909, 20mg/kg, BID, PO FHD-909, 40mg/kg, BID, PO FHD-909, 60mg/kg, BID, PO A549 Model RERF-LC-AI Model NCI-H1793 Model Genetic Background: SMARCA4, Q729fs / H736Y, KRAS G12S, STK11-/-, CDKN2A-/-, KEAP1 G333C Genetic Background: SMARCA4 mut p.E1496*, TP53 p.Q104*, NF1 p.E1699* Genetic Background: SMARCA4, E514*, TP53 R209* R273H, ARID1A C884* NOTE: All doses were well tolerated. Dosing holidays were applied to the 60 mg/kg dose groups, as appropriate.
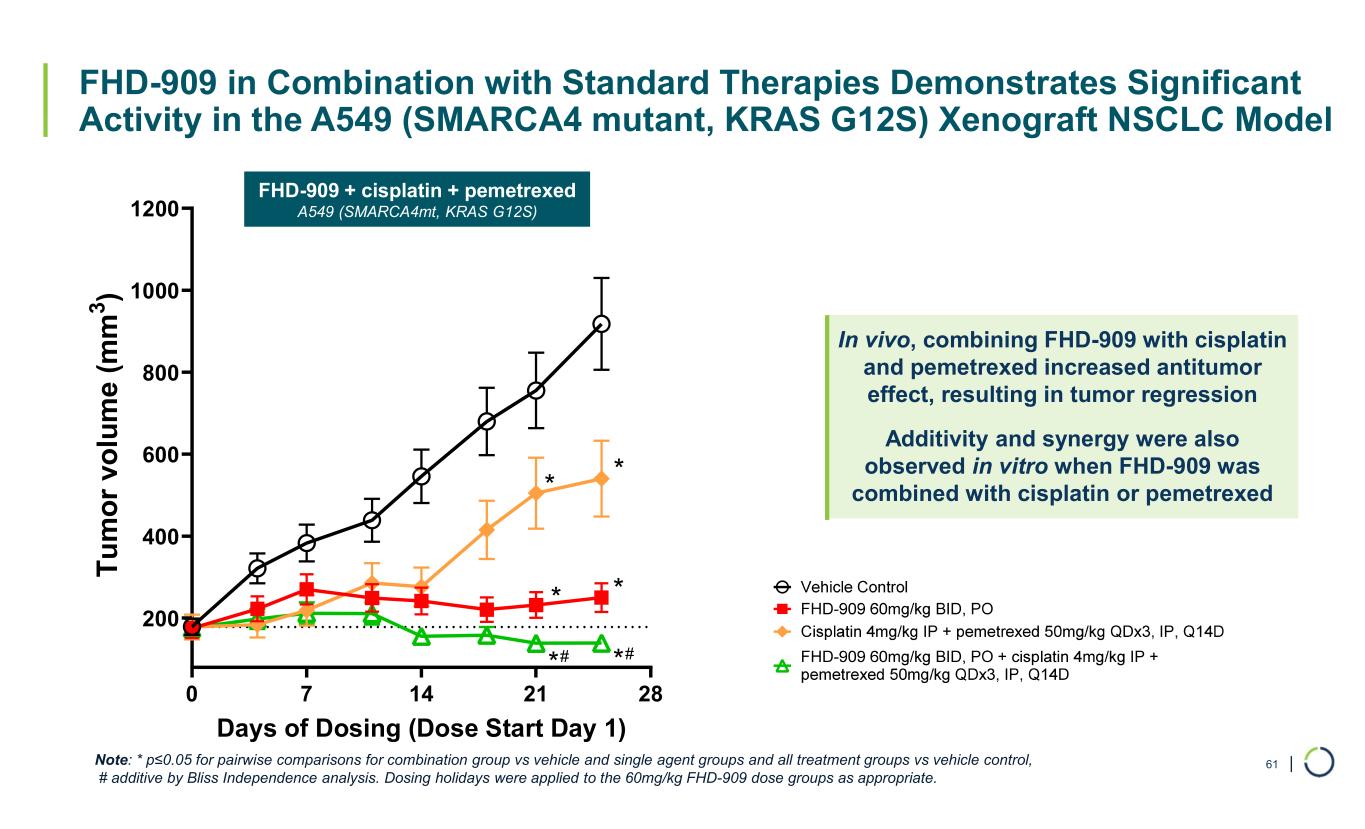
| FHD-909 in Combination with Standard Therapies Demonstrates Significant Activity in the A549 (SMARCA4 mutant, KRAS G12S) Xenograft NSCLC Model 61 0 7 14 21 28 200 400 600 800 1000 1200 Days of Dosing (Dose Start Day 1) T u m o r vo lu m e (m m 3 ) * * * * ** ## Note: * p≤0.05 for pairwise comparisons for combination group vs vehicle and single agent groups and all treatment groups vs vehicle control, # additive by Bliss Independence analysis. Dosing holidays were applied to the 60mg/kg FHD-909 dose groups as appropriate. FHD-909 + cisplatin + pemetrexed A549 (SMARCA4mt, KRAS G12S) In vivo, combining FHD-909 with cisplatin and pemetrexed increased antitumor effect, resulting in tumor regression Additivity and synergy were also observed in vitro when FHD-909 was combined with cisplatin or pemetrexed
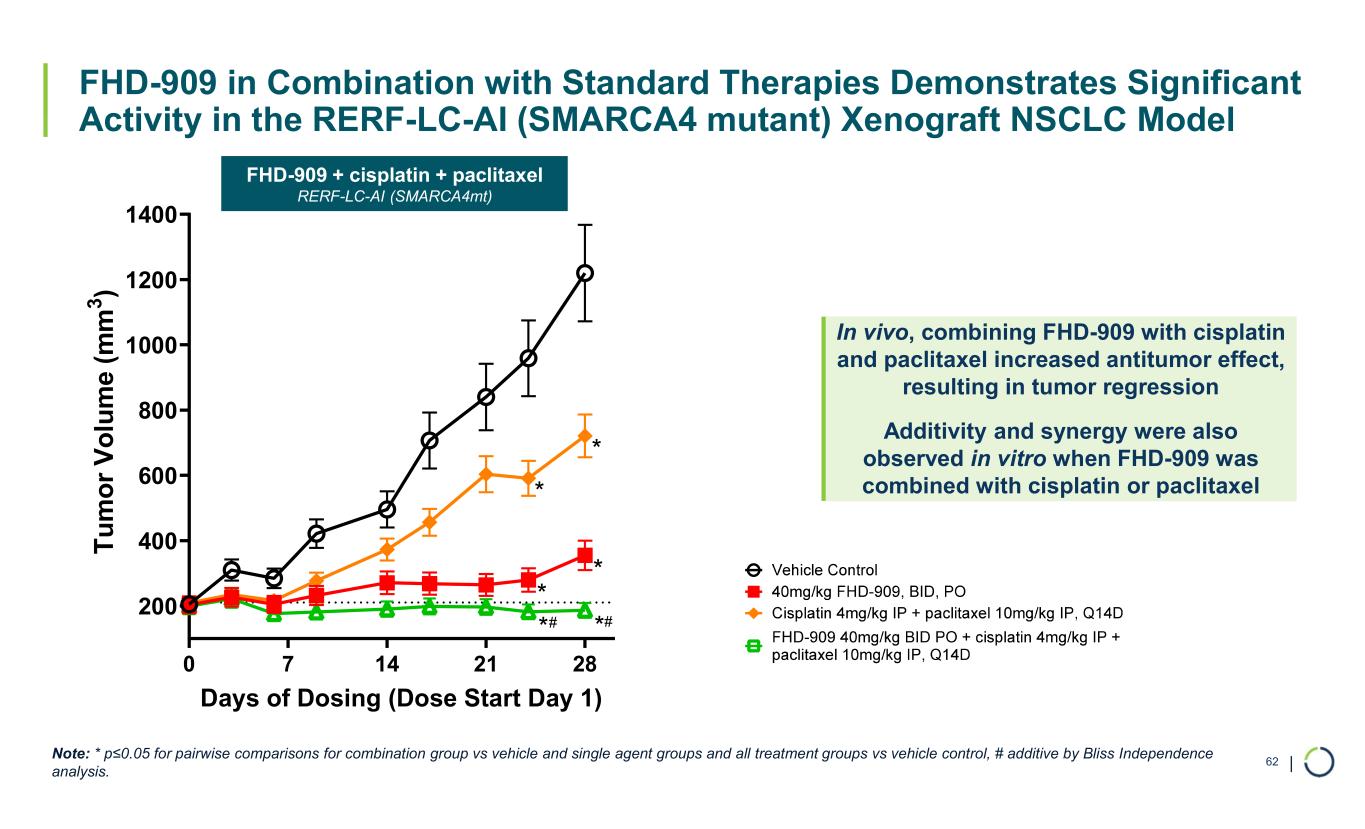
| FHD-909 in Combination with Standard Therapies Demonstrates Significant Activity in the RERF-LC-AI (SMARCA4 mutant) Xenograft NSCLC Model 62 0 7 14 21 28 200 400 600 800 1000 1200 1400 Days of Dosing (Dose Start Day 1) T u m o r V o lu m e (m m 3 ) * * * * **# # Note: * p≤0.05 for pairwise comparisons for combination group vs vehicle and single agent groups and all treatment groups vs vehicle control, # additive by Bliss Independence analysis. In vivo, combining FHD-909 with cisplatin and paclitaxel increased antitumor effect, resulting in tumor regression Additivity and synergy were also observed in vitro when FHD-909 was combined with cisplatin or paclitaxel FHD-909 + cisplatin + paclitaxel RERF-LC-AI (SMARCA4mt)

| Synergistic Activity Observed for FHD-909 in Combination with KRAS Inhibitors In Vitro 63 In h ib it io n ( % ) In h ib it io n ( % ) FHD-909 + olomorasib (KRAS G12C inhibitor) FHD-909 + pan-KRAS inhibitor In h ib it io n ( % ) In h ib it io n ( % ) 1 10 100 1000 10000 -20 0 20 40 60 80 100 120 FHD-909 concentration (nM) DMSO 7 14 28 56 113 225 450 900 [LY4066434](nM) In h ib it io n ( % ) NCI-H2030, Lung, NSCLC SMARCA4-/- KRAS(G12C) PATU-8988T, Pancreas, PDAC SMARCA4 - / fusion, KRAS(G12D)* SNU-407, Colon, CRC SMARCA4 P109fs194, R466C, Y507H, M1109V, KRAS(G12D) A549, Lung, NSCLC SMARCA4 Q729fs/H736Y, KRAS(G12S) PATU-8988T, Pancreas, PDAC SMARCA4 - / fusion, KRAS(G12V) Note: FHD-909 is reported in unbound concentrations in the assays; *CRISPR KI, fs frameshift FHD-909 + LY3962673 (KRAS G12D inhibitor) FHD-909 + LY4066434 (pan-KRAS inhibitor) FHD-909 + LY3962673 (KRAS G12D inhibitor) FHD-909 + LY4066434 (pan-KRAS inhibitor) NCI-H2030, Lung, NSCLC SMARCA4-/- KRAS(G12C)
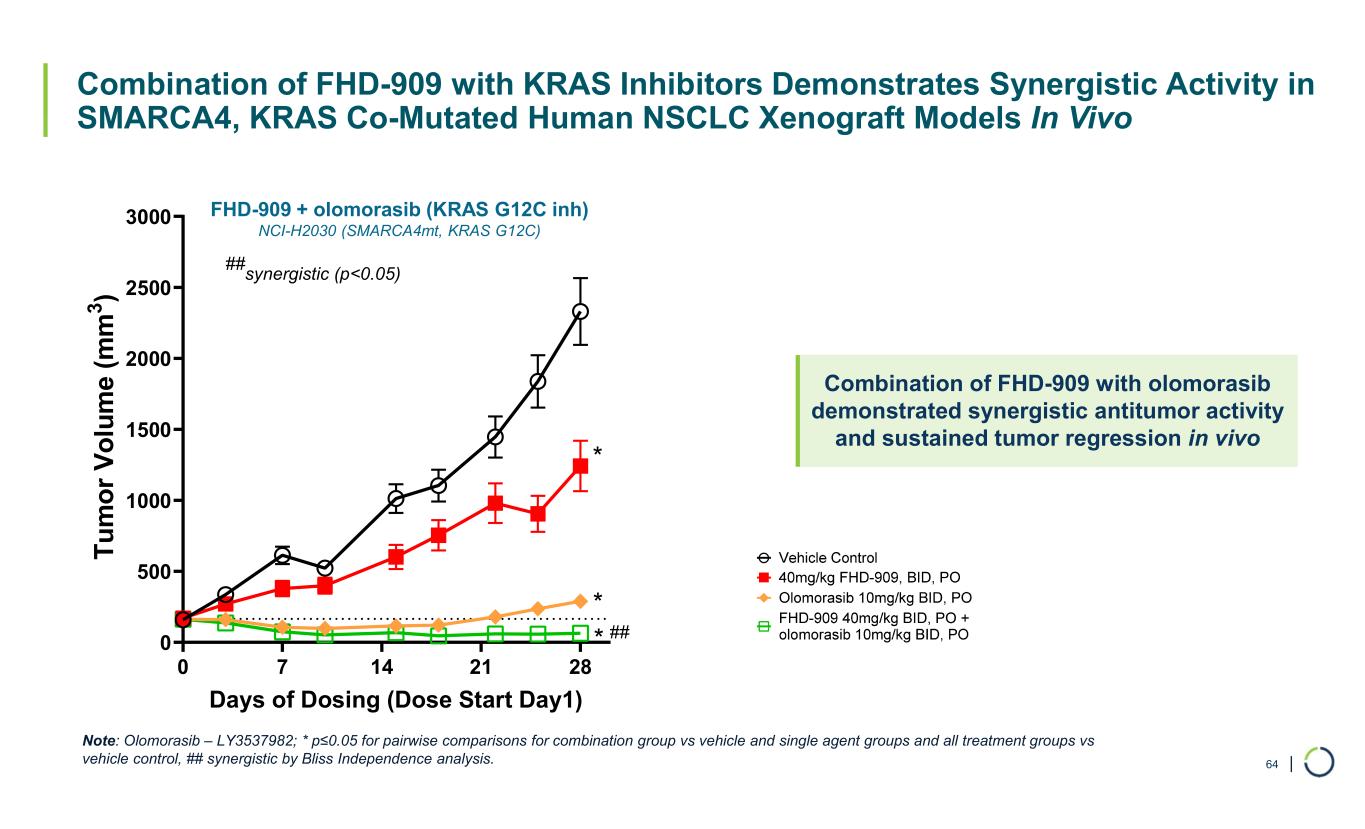
| Combination of FHD-909 with KRAS Inhibitors Demonstrates Synergistic Activity in SMARCA4, KRAS Co-Mutated Human NSCLC Xenograft Models In Vivo FHD-909 + olomorasib (KRAS G12C inh) NCI-H2030 (SMARCA4mt, KRAS G12C) 64 0 7 14 21 28 0 500 1000 1500 2000 2500 3000 Days of Dosing (Dose Start Day1) T u m o r V o lu m e (m m 3 ) ## synergistic (p<0.05) * * * ## Combination of FHD-909 with olomorasib demonstrated synergistic antitumor activity and sustained tumor regression in vivo Note: Olomorasib – LY3537982; * p≤0.05 for pairwise comparisons for combination group vs vehicle and single agent groups and all treatment groups vs vehicle control, ## synergistic by Bliss Independence analysis.
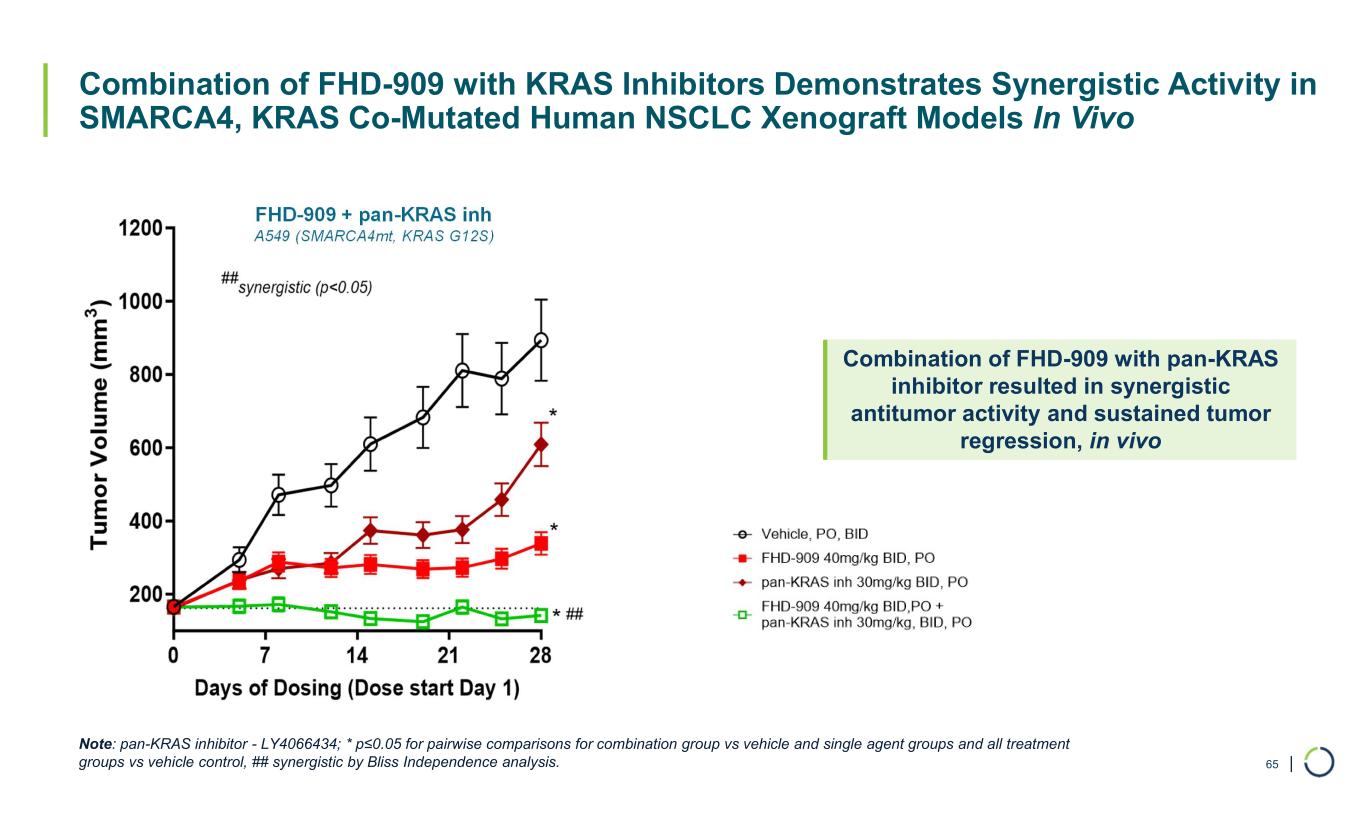
| Combination of FHD-909 with KRAS Inhibitors Demonstrates Synergistic Activity in SMARCA4, KRAS Co-Mutated Human NSCLC Xenograft Models In Vivo FHD-909 + pan-KRAS inh A549 (SMARCA4mt, KRAS G12S) 65 Combination of FHD-909 with pan-KRAS inhibitor resulted in synergistic antitumor activity and sustained tumor regression, in vivo Note: pan-KRAS inhibitor - LY4066434; * p≤0.05 for pairwise comparisons for combination group vs vehicle and single agent groups and all treatment groups vs vehicle control, ## synergistic by Bliss Independence analysis.

| FHD-909 in Combination with Pembrolizumab Shows Significantly Enhanced Anti-Tumor Activity in A549 CD34+ HSC Humanized Xenograft NSCLC Model 66 FHD-909 sensitized the tumor cells to pembrolizumab treatment resulting in enhanced combination activity. Pembrolizumab alone had no effect on tumor growth compared to vehicle control. Note: HSC, hematopoietic stem cells; * p≤0.05 for pairwise comparisons for combination group vs vehicle and single agents; # additive, ## synergistic by Bliss Independence analysis. FHD-909 (20 mg/kg) + pembrolizumab A549 (SMARCA4mt, KRAS G12S) FHD-909 (40 mg/kg) + pembrolizumab A549 (SMARCA4mt, KRAS G12S)
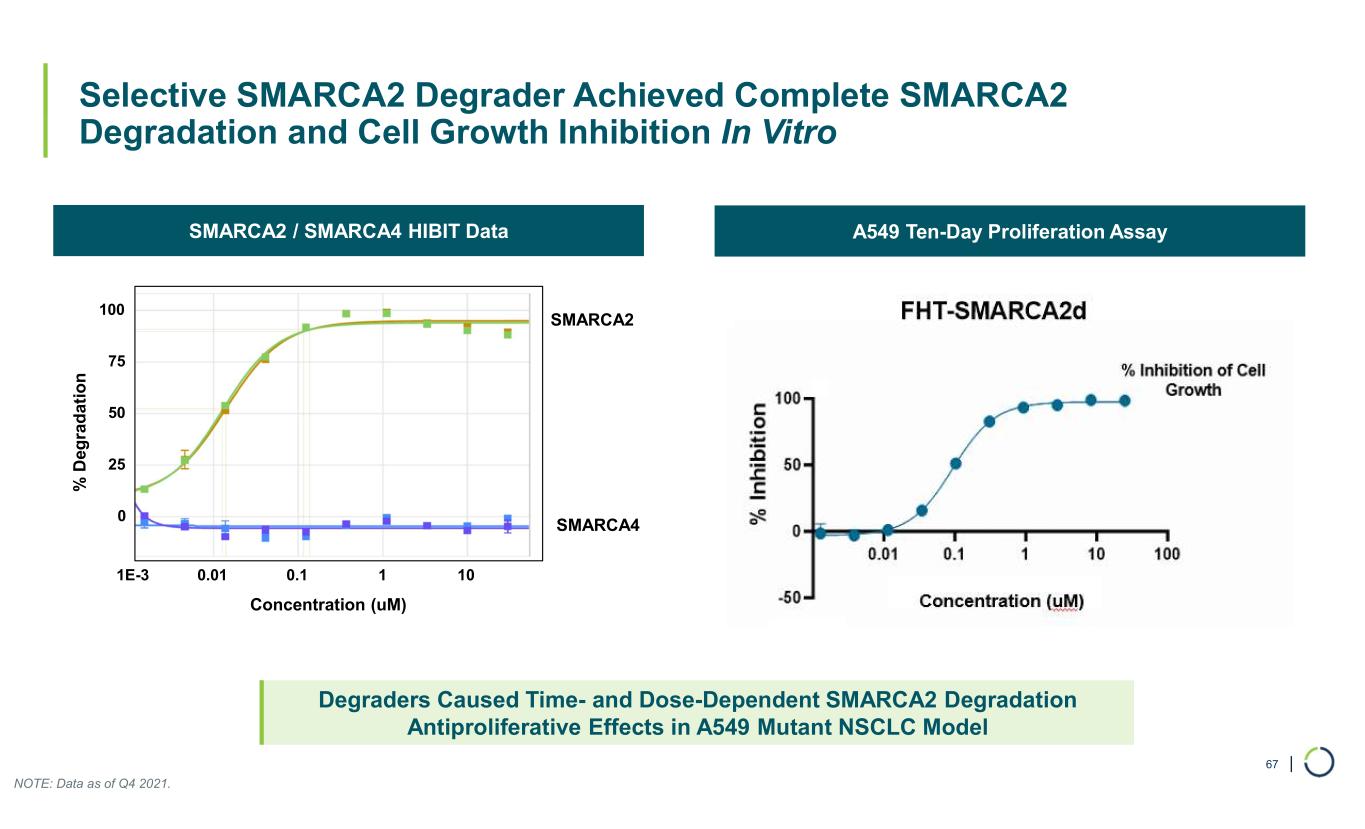
| Selective SMARCA2 Degrader Achieved Complete SMARCA2 Degradation and Cell Growth Inhibition In Vitro 67 Degraders Caused Time- and Dose-Dependent SMARCA2 Degradation Antiproliferative Effects in A549 Mutant NSCLC Model A549 Ten-Day Proliferation AssaySMARCA2 / SMARCA4 HIBIT Data SMARCA2 SMARCA4 % D eg ra d at io n Concentration (uM) 0 25 50 75 100 1E-3 0.01 0.1 1 10 NOTE: Data as of Q4 2021.

Protein Degradation Importance of Rate Analysis FHD-609 is a Selective, Potent, Protein Degrader of the BRD9 component of the BAF complex 68
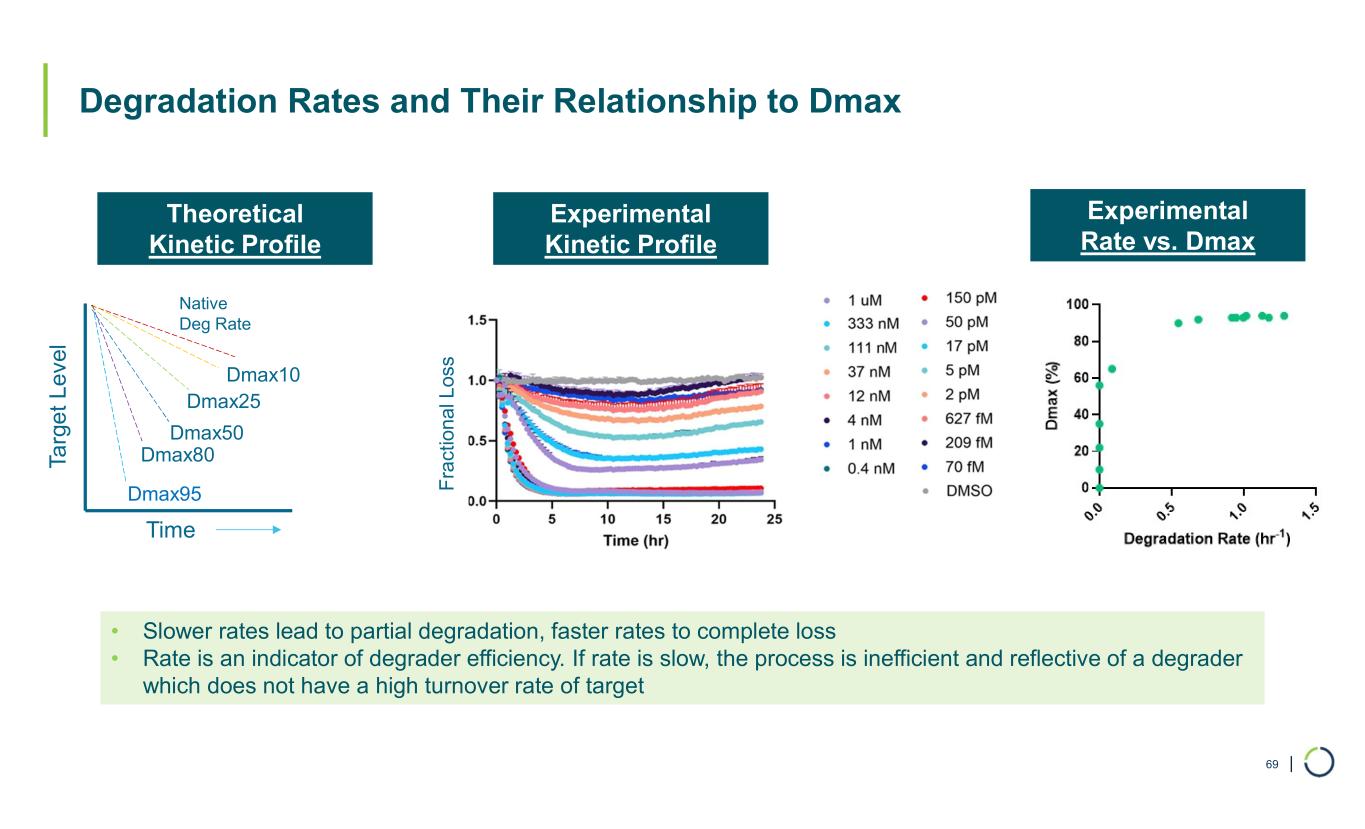
| Degradation Rates and Their Relationship to Dmax 69 • Slower rates lead to partial degradation, faster rates to complete loss • Rate is an indicator of degrader efficiency. If rate is slow, the process is inefficient and reflective of a degrader which does not have a high turnover rate of target Native Deg Rate Ta rg e t L e ve l Time Dmax10 Dmax25 Dmax50 Dmax80 Dmax95 Experimental Rate vs. Dmax Experimental Kinetic Profile Theoretical Kinetic Profile F ra ct io na l L os s

| Degradation Rate Dictates Dmax – Program Independent 70 Foghorn CBP ProgramFoghorn BRD9 Program Foghorn EP300 Program • Large scale experimental kinetic analysis for a program reveals the relationship between rate and Dmax
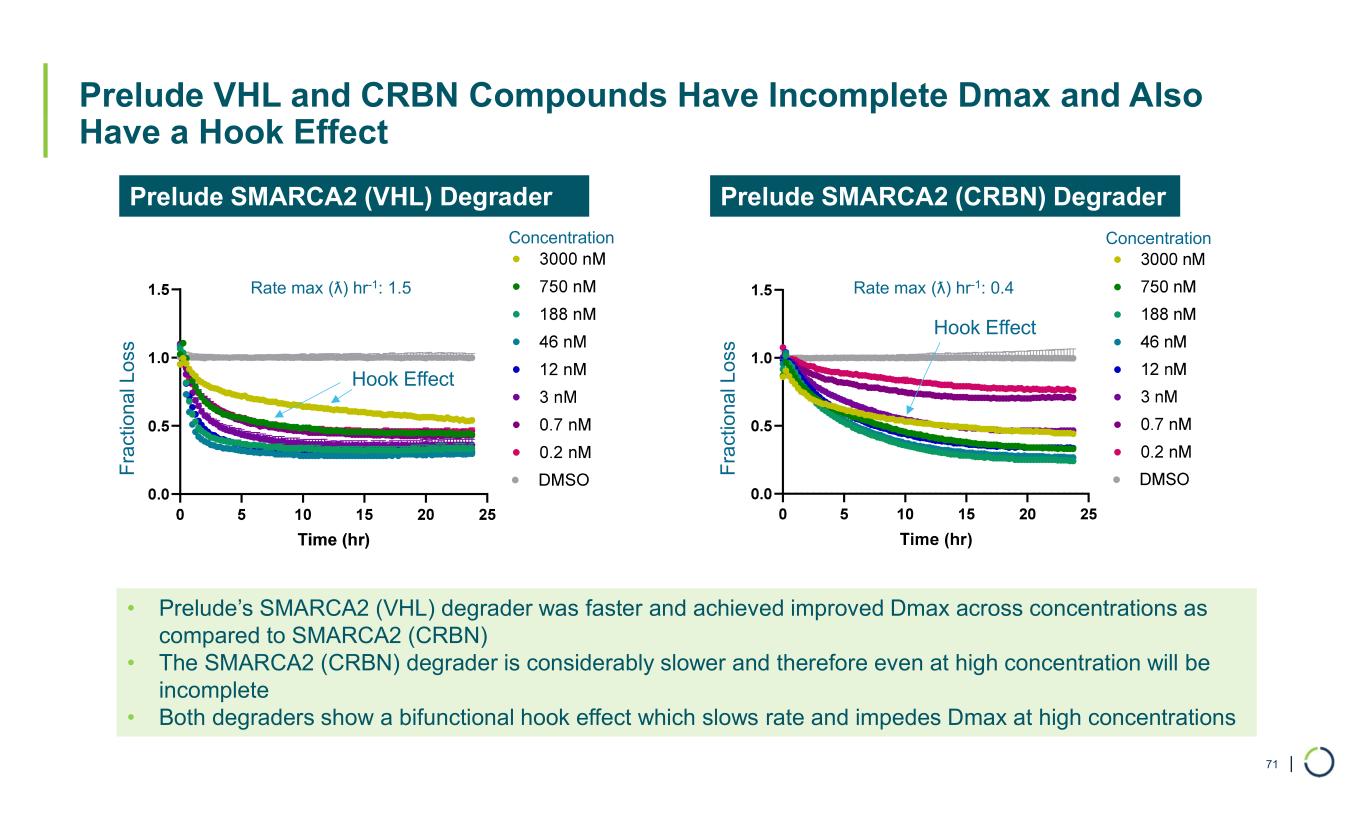
| Prelude VHL and CRBN Compounds Have Incomplete Dmax and Also Have a Hook Effect 71 • Prelude’s SMARCA2 (VHL) degrader was faster and achieved improved Dmax across concentrations as compared to SMARCA2 (CRBN) • The SMARCA2 (CRBN) degrader is considerably slower and therefore even at high concentration will be incomplete • Both degraders show a bifunctional hook effect which slows rate and impedes Dmax at high concentrations Prelude SMARCA2 (CRBN) DegraderPrelude SMARCA2 (VHL) Degrader F ra c ti o n a l L u m in e s ce n c e (R L U ) Concentration F ra ct io n a l L u m in es ce n ce (R L U ) Concentration Hook Effect Hook Effect Rate max (ƛ) hr-1: 0.4Rate max (ƛ) hr-1: 1.5 F ra ct io na l L os s F ra ct io na l L os s
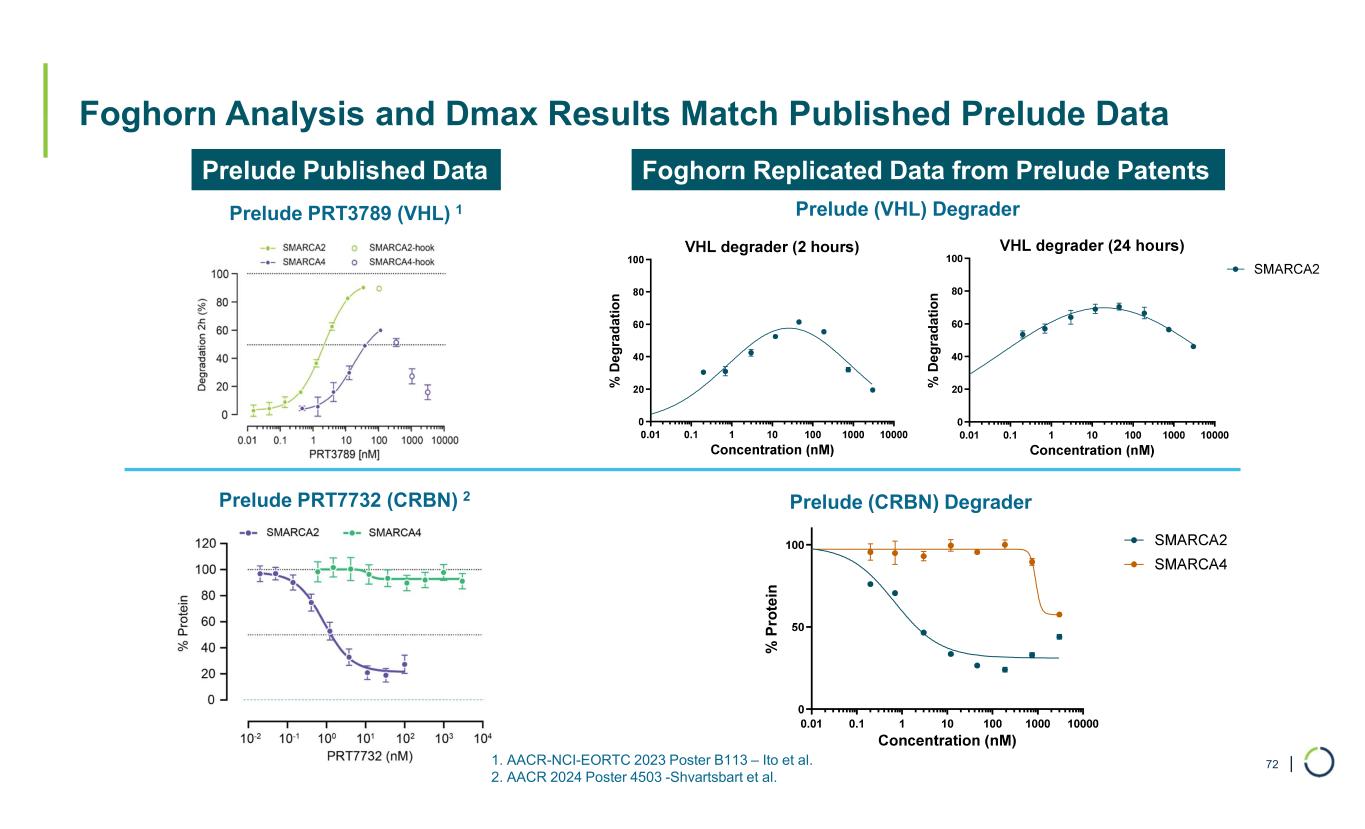
| Foghorn Analysis and Dmax Results Match Published Prelude Data 72 Prelude Published Data Prelude PRT3789 (VHL) 1 Prelude PRT7732 (CRBN) 2 % P ro te in Prelude (CRBN) Degrader Prelude (VHL) Degrader Foghorn Replicated Data from Prelude Patents 1. AACR-NCI-EORTC 2023 Poster B113 – Ito et al. 2. AACR 2024 Poster 4503 -Shvartsbart et al. % D e g ra d at io n % D e g ra d a ti o n

Induced Proximity Platform Evolution FHD-609 is a Selective, Potent, Protein Degrader of the BRD9 component of the BAF complex 73

| Existing Capabilities Have Produced Multiple Degrader Programs and are Setting Foundation for Expansion Into Induced Proximity 74 TargetDegrader Machinery (E3 Ligase) TargetEffector Post-Translational modification - dependent Protein Stabilization Modulation of gene activation/repression Induced Proximity describes a chemical means to bring together a target and effector to re-program disease states Targeted protein degradation is a compelling example of induced proximity Induced proximity offers an almost endless opportunity to modulate biology
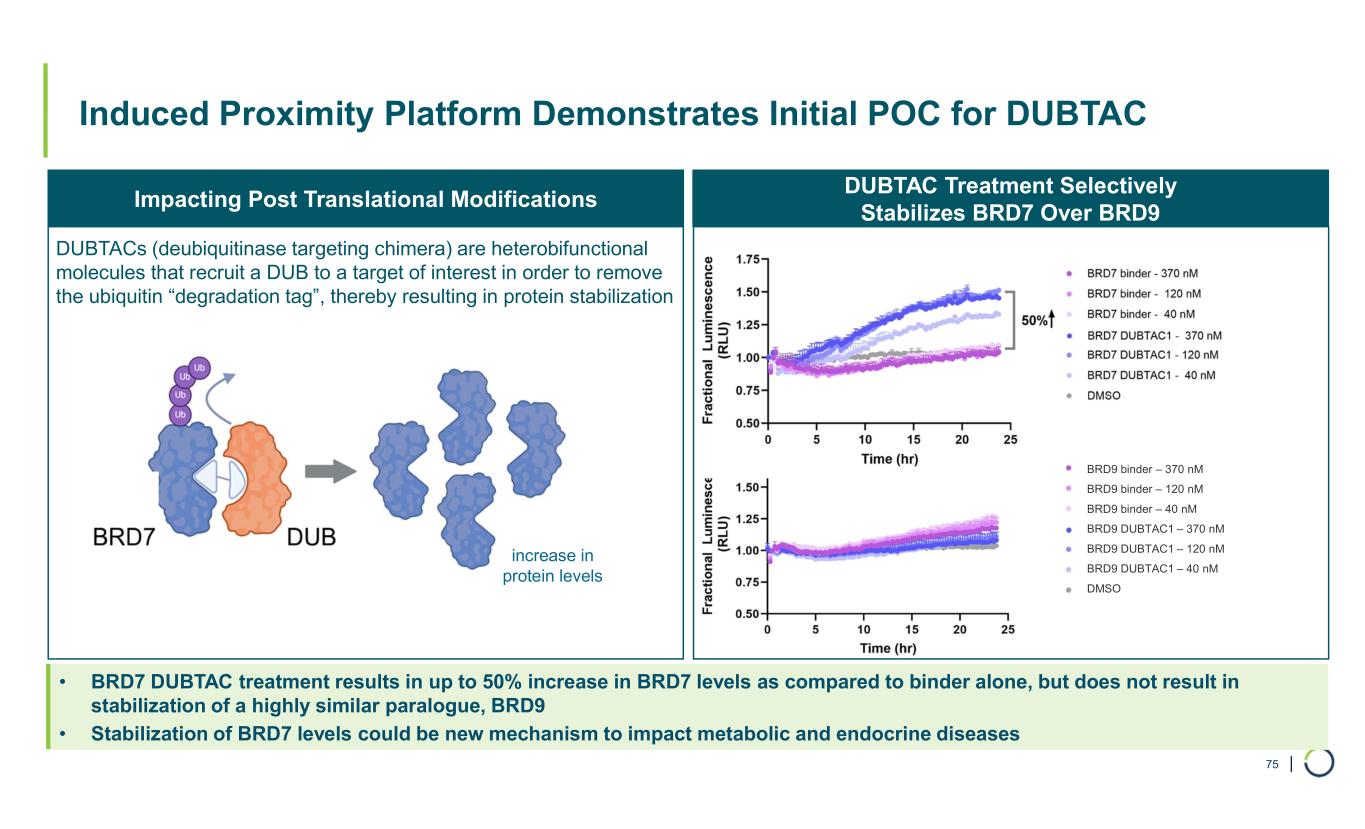
| Induced Proximity Platform Demonstrates Initial POC for DUBTAC 75 increase in protein levels DUBTACs (deubiquitinase targeting chimera) are heterobifunctional molecules that recruit a DUB to a target of interest in order to remove the ubiquitin “degradation tag”, thereby resulting in protein stabilization DUBTAC Treatment Selectively Stabilizes BRD7 Over BRD9 • BRD7 DUBTAC treatment results in up to 50% increase in BRD7 levels as compared to binder alone, but does not result in stabilization of a highly similar paralogue, BRD9 • Stabilization of BRD7 levels could be new mechanism to impact metabolic and endocrine diseases Impacting Post Translational Modifications BRD9 binder – 370 nM BRD9 binder – 120 nM BRD9 binder – 40 nM BRD9 DUBTAC1 – 370 nM BRD9 DUBTAC1 – 120 nM BRD9 DUBTAC1 – 40 nM DMSO

| Induced Proximity Platform Demonstrates Potential for Novel Approach to Activate Cell Death Cascade 76 Normal BCL6 Function BCL6 Activation with BAF-BCL6 Heterobifunctional • BCL6 is a proto-oncogene, which binds to promoters of cell death genes and suppresses their expression • We have developed a hetero-bifunctional molecule which couples the chromatin remodeling complex BAF with BCL6 • BAF releases the BCL6 complex, which activates the cell death cascade killing tumor cells • BCL6 inhibitors: GNE and BI, administered on their own have no effect on tumor cells • Genes downstream of BCL6 become more highly expressed when coupled with BAF vs. the BCL6 inhibitor alone STOP DMSO GNE (EC50: 81.65 μM) BI (EC50: 24.34 μM) FHTX (EC50: 0.8614 μM) GNE + BI (EC50: 16.16 μM) KARPAS422 Modulating Gene Activation/Repression BAF-BCL6 Heterobifunctional Releases Cell Death Cascade

| 48 Nuclear Hormone Receptors Nuclear hormone receptor are implicated in multiple biology processes across most organ systems • Only 10 nuclear hormone receptors have been drugged and approved to date (e.g., AR, ER Glucocorticoids, thyroid HR alpha beta) • Despite decades of development most others have not successfully been developed into drugs for various reasons (e.g. lack of efficacy, safety) • Make existing agonists or antagonists more efficacious • Improve upon failed approaches by addressing efficacy and/or safety issue ~ 10 Nuclear Hormone Receptors Applications of FHTX Induced Proximity Platfrom Access to multiple TAs and indications given promiscuity of HR across multitude of biology processes Induced Proximity Platform Offers Expansion Beyond Oncology; Nuclear Hormone Receptors Initial Area of Focus – Potential for Partnerships 77
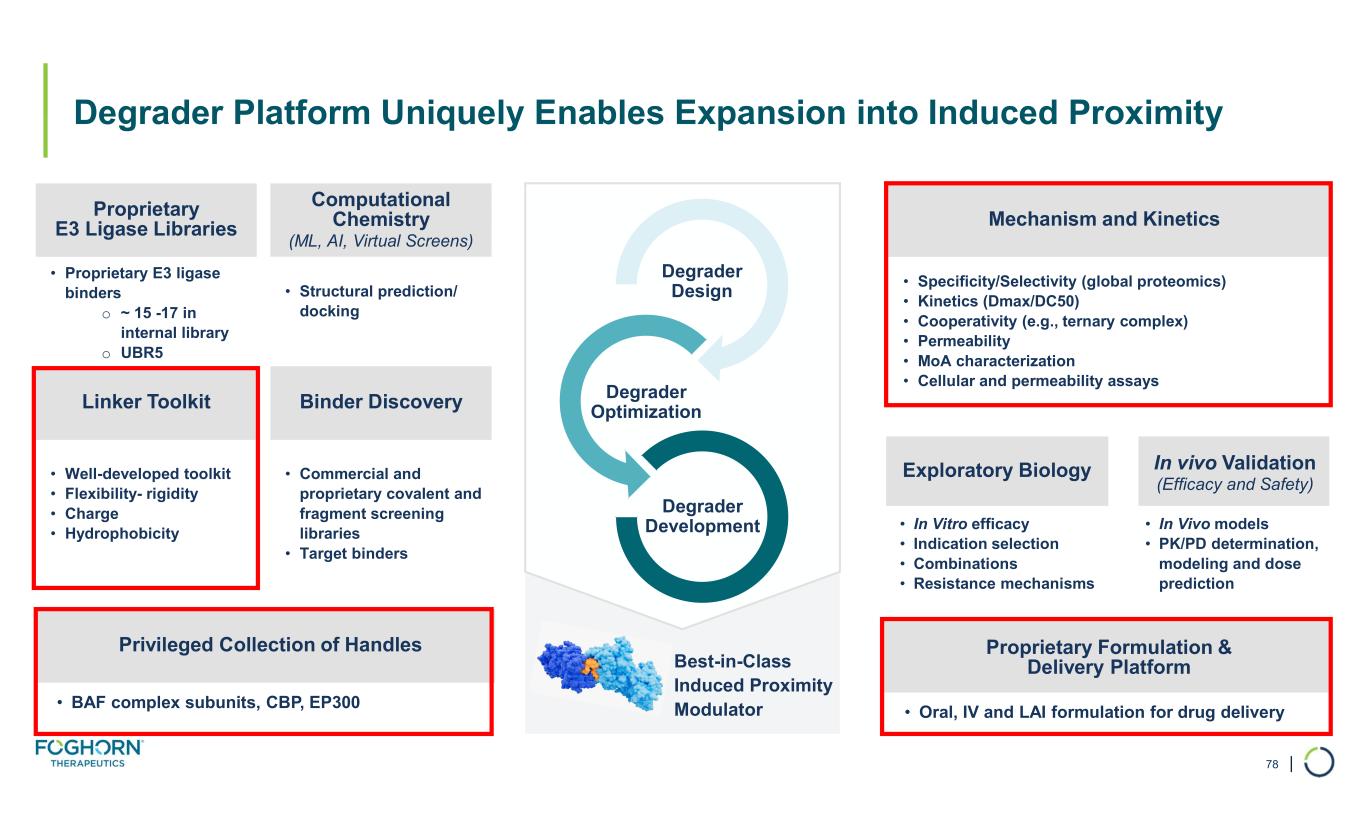
| Degrader Design Degrader Optimization Degrader Development Best-in-Class Induced Proximity Modulator • Structural prediction/ docking • Well-developed toolkit • Flexibility- rigidity • Charge • Hydrophobicity • Commercial and proprietary covalent and fragment screening libraries • Target binders • Oral, IV and LAI formulation for drug delivery Computational Chemistry (ML, AI, Virtual Screens) Proprietary E3 Ligase Libraries Binder DiscoveryLinker Toolkit Mechanism and Kinetics Exploratory Biology In vivo Validation (Efficacy and Safety) Proprietary Formulation & Delivery Platform • BAF complex subunits, CBP, EP300 Privileged Collection of Handles Degrader Platform Uniquely Enables Expansion into Induced Proximity 78 • Proprietary E3 ligase binders o ~ 15 -17 in internal library o UBR5 • Specificity/Selectivity (global proteomics) • Kinetics (Dmax/DC50) • Cooperativity (e.g., ternary complex) • Permeability • MoA characterization • Cellular and permeability assays • In Vitro efficacy • Indication selection • Combinations • Resistance mechanisms • In Vivo models • PK/PD determination, modeling and dose prediction

Drugging Transcription Factors Multiple Approaches FHD-609 is a Selective, Potent, Protein Degrader of the BRD9 component of the BAF complex 79
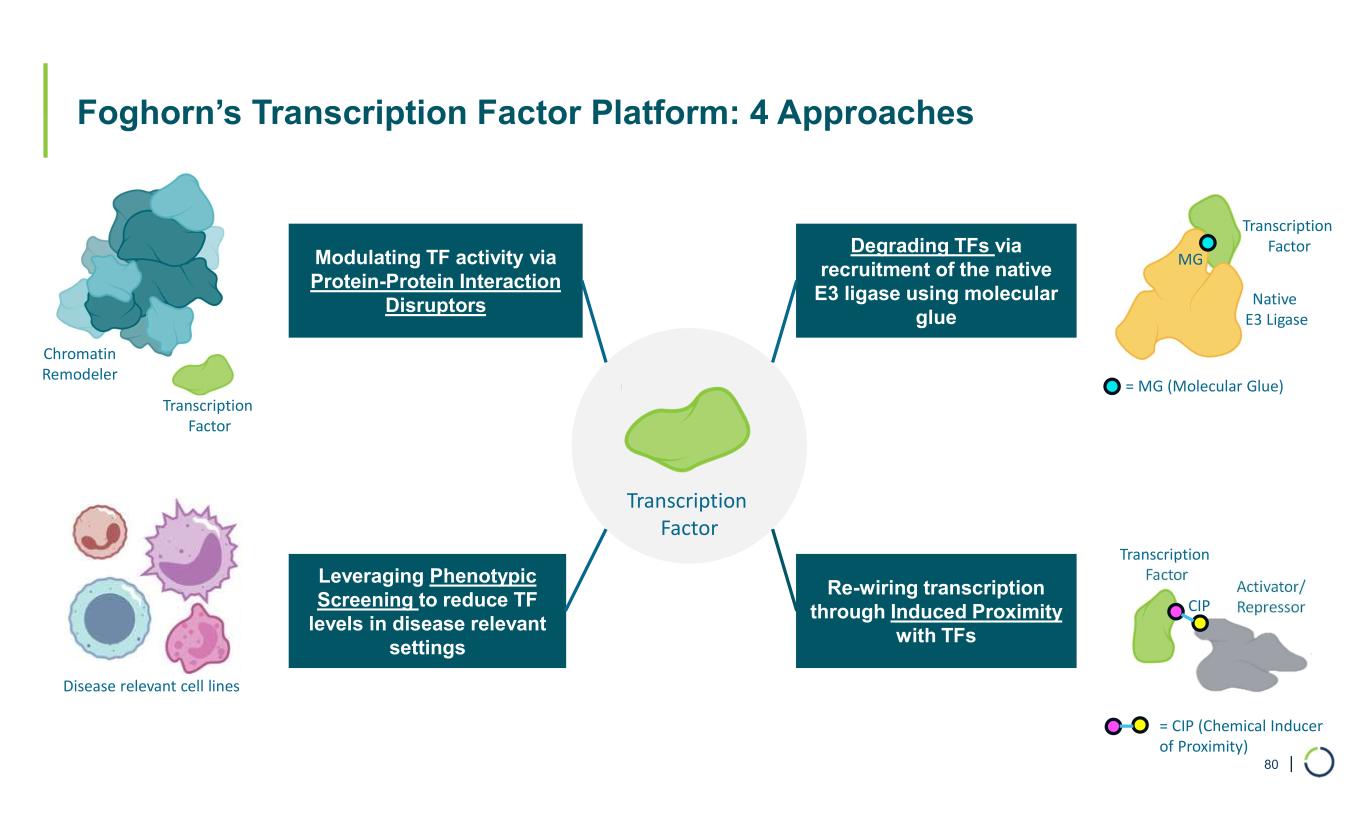
| Foghorn’s Transcription Factor Platform: 4 Approaches 80 Transcription Factor Chromatin Remodeler Modulating TF activity via Protein-Protein Interaction Disruptors Transcription Factor Native E3 Ligase Degrading TFs via recruitment of the native E3 ligase using molecular glue = MG (Molecular Glue) Leveraging Phenotypic Screening to reduce TF levels in disease relevant settings Re-wiring transcription through Induced Proximity with TFs Disease relevant cell lines Transcription Factor Activator/ Repressor = CIP (Chemical Inducer of Proximity) CIP MG Transcription Factor















































































